| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
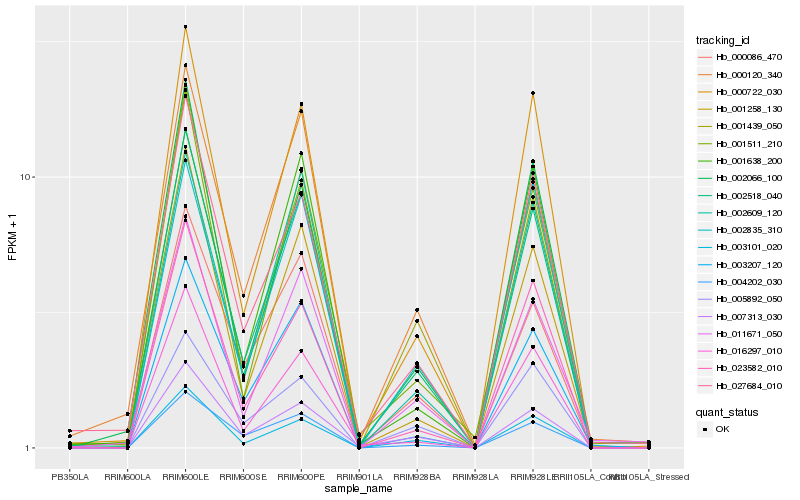
Hb_002518_040 |
0.0 |
- |
- |
PREDICTED: probable serine/threonine-protein kinase At5g41260 [Jatropha curcas] |
| 2 |
Hb_000722_030 |
0.0499931015 |
- |
- |
putative plant disease resistance family protein [Populus trichocarpa] |
| 3 |
Hb_003101_020 |
0.075589492 |
transcription factor |
TF Family: NAC |
PREDICTED: NAC transcription factor NAM-B2 [Jatropha curcas] |
| 4 |
Hb_001439_050 |
0.0808019427 |
- |
- |
plastocyanin-like domain-containing family protein [Populus trichocarpa] |
| 5 |
Hb_001258_130 |
0.0810776862 |
- |
- |
PREDICTED: kinesin-like protein KIF19 [Jatropha curcas] |
| 6 |
Hb_007313_030 |
0.0830383614 |
- |
- |
PREDICTED: receptor-like cytosolic serine/threonine-protein kinase RBK2 [Jatropha curcas] |
| 7 |
Hb_027684_010 |
0.0844805644 |
- |
- |
hypothetical protein RCOM_0504250 [Ricinus communis] |
| 8 |
Hb_023582_010 |
0.0857658984 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 9 |
Hb_003207_120 |
0.0869502292 |
- |
- |
molybdopterin cofactor sulfurase, putative [Ricinus communis] |
| 10 |
Hb_002609_120 |
0.0877283824 |
- |
- |
hypothetical protein POPTR_0001s25660g [Populus trichocarpa] |
| 11 |
Hb_011671_050 |
0.0884657069 |
- |
- |
PREDICTED: F-box protein At5g07670 [Jatropha curcas] |
| 12 |
Hb_000086_470 |
0.089100106 |
- |
- |
hypothetical protein JCGZ_17645 [Jatropha curcas] |
| 13 |
Hb_001511_210 |
0.0895513088 |
- |
- |
PREDICTED: protein ESKIMO 1-like [Jatropha curcas] |
| 14 |
Hb_016297_010 |
0.0900904853 |
- |
- |
PREDICTED: uncharacterized protein LOC105638665 [Jatropha curcas] |
| 15 |
Hb_002066_100 |
0.0941956981 |
- |
- |
PREDICTED: protein disulfide-isomerase SCO2 [Jatropha curcas] |
| 16 |
Hb_005892_050 |
0.095110326 |
- |
- |
PREDICTED: uncharacterized protein LOC105632175 [Jatropha curcas] |
| 17 |
Hb_000120_340 |
0.097774479 |
- |
- |
Cucumisin precursor, putative [Ricinus communis] |
| 18 |
Hb_002835_310 |
0.100457709 |
- |
- |
ATP binding protein, putative [Ricinus communis] |
| 19 |
Hb_001638_200 |
0.1017120492 |
transcription factor |
TF Family: NAC |
PREDICTED: NAC domain-containing protein 43 [Jatropha curcas] |
| 20 |
Hb_004202_030 |
0.101841633 |
- |
- |
PREDICTED: uncharacterized protein LOC103449702 [Malus domestica] |