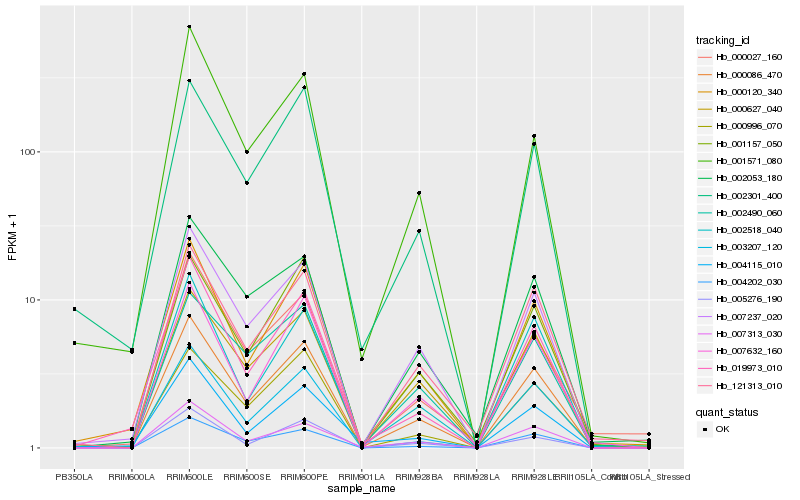
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000086_470 |
0.0 |
- |
- |
hypothetical protein JCGZ_17645 [Jatropha curcas] |
| 2 |
Hb_007237_020 |
0.0630450599 |
- |
- |
PREDICTED: protein trichome birefringence-like 23 [Jatropha curcas] |
| 3 |
Hb_000120_340 |
0.0704013781 |
- |
- |
Cucumisin precursor, putative [Ricinus communis] |
| 4 |
Hb_004202_030 |
0.0719199355 |
- |
- |
PREDICTED: uncharacterized protein LOC103449702 [Malus domestica] |
| 5 |
Hb_007313_030 |
0.0842869216 |
- |
- |
PREDICTED: receptor-like cytosolic serine/threonine-protein kinase RBK2 [Jatropha curcas] |
| 6 |
Hb_007632_160 |
0.0848211858 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 7 |
Hb_000627_040 |
0.0851224444 |
- |
- |
leucine-rich repeat protein, putative [Ricinus communis] |
| 8 |
Hb_019973_010 |
0.0858864907 |
- |
- |
PREDICTED: probable methyltransferase PMT18 isoform X2 [Jatropha curcas] |
| 9 |
Hb_121313_010 |
0.0884547073 |
- |
- |
PREDICTED: endoglucanase 24-like [Jatropha curcas] |
| 10 |
Hb_002518_040 |
0.089100106 |
- |
- |
PREDICTED: probable serine/threonine-protein kinase At5g41260 [Jatropha curcas] |
| 11 |
Hb_001157_050 |
0.0917916739 |
- |
- |
cation efflux protein/ zinc transporter, putative [Ricinus communis] |
| 12 |
Hb_003207_120 |
0.0923118973 |
- |
- |
molybdopterin cofactor sulfurase, putative [Ricinus communis] |
| 13 |
Hb_002053_180 |
0.0992507845 |
- |
- |
PREDICTED: probable inactive receptor kinase At2g26730 [Jatropha curcas] |
| 14 |
Hb_002490_060 |
0.101002955 |
- |
- |
PREDICTED: uncharacterized protein LOC105638743 [Jatropha curcas] |
| 15 |
Hb_000996_070 |
0.1054752127 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 16 |
Hb_000027_160 |
0.1072018638 |
- |
- |
PREDICTED: probable protein phosphatase 2C 15 isoform X3 [Jatropha curcas] |
| 17 |
Hb_001571_080 |
0.1075528139 |
- |
- |
PREDICTED: non-specific lipid-transfer protein-like protein At5g64080 [Jatropha curcas] |
| 18 |
Hb_004115_010 |
0.109715068 |
transcription factor |
TF Family: HB |
PREDICTED: homeobox-leucine zipper protein HDG11 [Jatropha curcas] |
| 19 |
Hb_005276_190 |
0.1099720244 |
- |
- |
PREDICTED: uncharacterized protein LOC105650003 isoform X2 [Jatropha curcas] |
| 20 |
Hb_002301_400 |
0.11159629 |
- |
- |
structural constituent of cell wall, putative [Ricinus communis] |