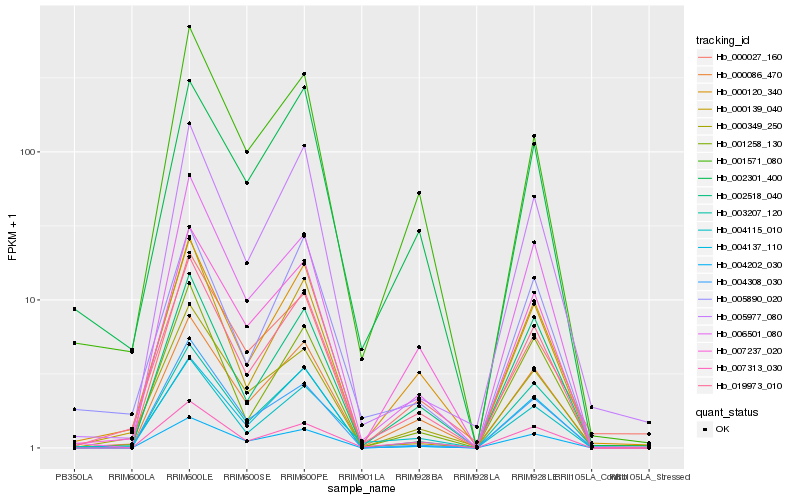
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_019973_010 |
0.0 |
- |
- |
PREDICTED: probable methyltransferase PMT18 isoform X2 [Jatropha curcas] |
| 2 |
Hb_000120_340 |
0.0766117928 |
- |
- |
Cucumisin precursor, putative [Ricinus communis] |
| 3 |
Hb_004115_010 |
0.0773937766 |
transcription factor |
TF Family: HB |
PREDICTED: homeobox-leucine zipper protein HDG11 [Jatropha curcas] |
| 4 |
Hb_000086_470 |
0.0858864907 |
- |
- |
hypothetical protein JCGZ_17645 [Jatropha curcas] |
| 5 |
Hb_000349_250 |
0.087624905 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At1g12460 [Jatropha curcas] |
| 6 |
Hb_004202_030 |
0.0991832348 |
- |
- |
PREDICTED: uncharacterized protein LOC103449702 [Malus domestica] |
| 7 |
Hb_003207_120 |
0.1019614529 |
- |
- |
molybdopterin cofactor sulfurase, putative [Ricinus communis] |
| 8 |
Hb_000139_040 |
0.1040855786 |
- |
- |
spermine synthase, putative [Ricinus communis] |
| 9 |
Hb_001571_080 |
0.1048252 |
- |
- |
PREDICTED: non-specific lipid-transfer protein-like protein At5g64080 [Jatropha curcas] |
| 10 |
Hb_004137_110 |
0.1078968752 |
- |
- |
PREDICTED: probable auxin efflux carrier component 6 [Jatropha curcas] |
| 11 |
Hb_002518_040 |
0.1081107458 |
- |
- |
PREDICTED: probable serine/threonine-protein kinase At5g41260 [Jatropha curcas] |
| 12 |
Hb_001258_130 |
0.1087025278 |
- |
- |
PREDICTED: kinesin-like protein KIF19 [Jatropha curcas] |
| 13 |
Hb_006501_080 |
0.1089491114 |
- |
- |
PREDICTED: uncharacterized protein LOC105637775 [Jatropha curcas] |
| 14 |
Hb_007237_020 |
0.1092147306 |
- |
- |
PREDICTED: protein trichome birefringence-like 23 [Jatropha curcas] |
| 15 |
Hb_005977_080 |
0.1103934365 |
transcription factor |
TF Family: bZIP |
PREDICTED: transcription factor HY5-like [Jatropha curcas] |
| 16 |
Hb_007313_030 |
0.1168518529 |
- |
- |
PREDICTED: receptor-like cytosolic serine/threonine-protein kinase RBK2 [Jatropha curcas] |
| 17 |
Hb_005890_020 |
0.1181218726 |
- |
- |
putative plant disease resistance family protein [Populus trichocarpa] |
| 18 |
Hb_000027_160 |
0.1200074734 |
- |
- |
PREDICTED: probable protein phosphatase 2C 15 isoform X3 [Jatropha curcas] |
| 19 |
Hb_002301_400 |
0.1209988674 |
- |
- |
structural constituent of cell wall, putative [Ricinus communis] |
| 20 |
Hb_004308_030 |
0.1214897871 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At4g08850 [Jatropha curcas] |