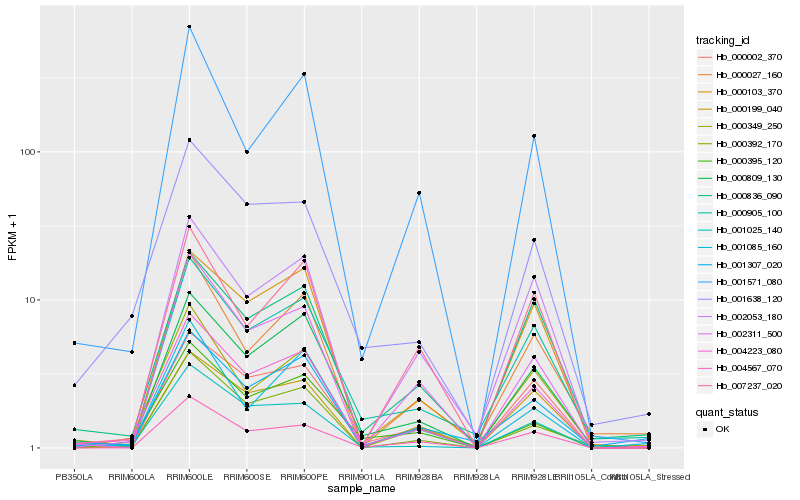
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_004223_080 |
0.0 |
transcription factor |
TF Family: C2H2 |
nucleic acid binding protein, putative [Ricinus communis] |
| 2 |
Hb_000809_130 |
0.0932243751 |
- |
- |
PREDICTED: peroxidase 10 isoform X3 [Jatropha curcas] |
| 3 |
Hb_000002_370 |
0.1044078661 |
- |
- |
PREDICTED: uncharacterized protein LOC105632103 [Jatropha curcas] |
| 4 |
Hb_001025_140 |
0.1045781703 |
- |
- |
hypothetical protein POPTR_0015s09180g [Populus trichocarpa] |
| 5 |
Hb_000392_170 |
0.1060682748 |
- |
- |
PREDICTED: protein PHYTOCHROME KINASE SUBSTRATE 3 [Jatropha curcas] |
| 6 |
Hb_000349_250 |
0.1079789539 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At1g12460 [Jatropha curcas] |
| 7 |
Hb_001307_020 |
0.1083460145 |
- |
- |
electron carrier, putative [Ricinus communis] |
| 8 |
Hb_002311_500 |
0.1107350678 |
- |
- |
PREDICTED: uncharacterized protein LOC105643461 [Jatropha curcas] |
| 9 |
Hb_001571_080 |
0.1118479859 |
- |
- |
PREDICTED: non-specific lipid-transfer protein-like protein At5g64080 [Jatropha curcas] |
| 10 |
Hb_004567_070 |
0.1175095631 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_001638_120 |
0.118180554 |
- |
- |
PREDICTED: CBL-interacting serine/threonine-protein kinase 9 [Jatropha curcas] |
| 12 |
Hb_000905_100 |
0.1195545268 |
transcription factor |
TF Family: bHLH |
PREDICTED: transcription factor bHLH130-like isoform X1 [Jatropha curcas] |
| 13 |
Hb_000836_090 |
0.1196766221 |
- |
- |
PREDICTED: calmodulin-binding receptor-like cytoplasmic kinase 2 [Jatropha curcas] |
| 14 |
Hb_000103_370 |
0.1201188515 |
- |
- |
PREDICTED: protein argonaute 10 [Jatropha curcas] |
| 15 |
Hb_002053_180 |
0.1202590687 |
- |
- |
PREDICTED: probable inactive receptor kinase At2g26730 [Jatropha curcas] |
| 16 |
Hb_000027_160 |
0.1211480741 |
- |
- |
PREDICTED: probable protein phosphatase 2C 15 isoform X3 [Jatropha curcas] |
| 17 |
Hb_000199_040 |
0.122029952 |
desease resistance |
Gene Name: TIR |
PREDICTED: TMV resistance protein N-like isoform X1 [Jatropha curcas] |
| 18 |
Hb_000395_120 |
0.1264568356 |
- |
- |
Stellacyanin, putative [Ricinus communis] |
| 19 |
Hb_001085_160 |
0.1329422782 |
- |
- |
PREDICTED: PLASMODESMATA CALLOSE-BINDING PROTEIN 3 [Jatropha curcas] |
| 20 |
Hb_007237_020 |
0.132967274 |
- |
- |
PREDICTED: protein trichome birefringence-like 23 [Jatropha curcas] |