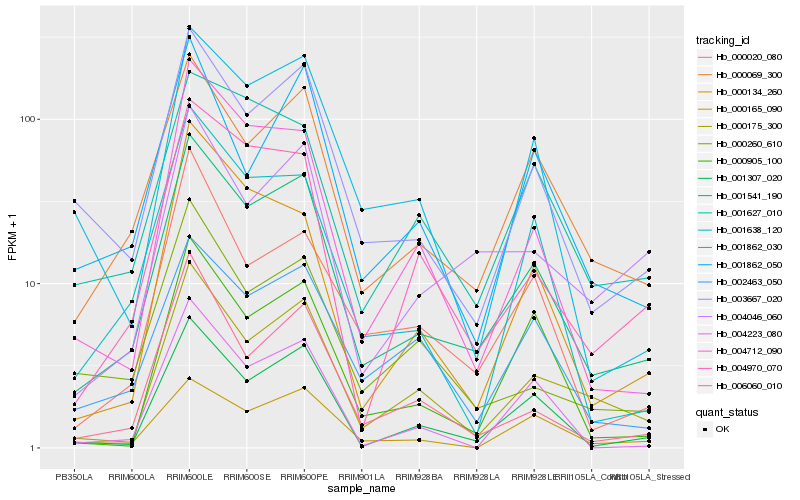
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001541_190 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105644700 [Jatropha curcas] |
| 2 |
Hb_000069_300 |
0.0991055313 |
- |
- |
Aspartic proteinase nepenthesin-1 precursor, putative [Ricinus communis] |
| 3 |
Hb_001307_020 |
0.1096909856 |
- |
- |
electron carrier, putative [Ricinus communis] |
| 4 |
Hb_003667_020 |
0.1120048924 |
- |
- |
conserved hypothetical protein 16 [Hevea brasiliensis] |
| 5 |
Hb_000175_300 |
0.1267595565 |
- |
- |
PREDICTED: uncharacterized protein LOC105634858 [Jatropha curcas] |
| 6 |
Hb_001638_120 |
0.1387878885 |
- |
- |
PREDICTED: CBL-interacting serine/threonine-protein kinase 9 [Jatropha curcas] |
| 7 |
Hb_004970_070 |
0.1393988102 |
- |
- |
hypothetical protein POPTR_0002s12620g [Populus trichocarpa] |
| 8 |
Hb_004712_090 |
0.1412580555 |
- |
- |
Serine acetyltransferase 3, mitochondrial precursor, putative [Ricinus communis] |
| 9 |
Hb_006060_010 |
0.1449424445 |
- |
- |
PREDICTED: cyclin-A2-2-like [Jatropha curcas] |
| 10 |
Hb_001862_030 |
0.1458757068 |
- |
- |
PREDICTED: probable esterase KAI2 [Jatropha curcas] |
| 11 |
Hb_000165_090 |
0.1475415693 |
- |
- |
PREDICTED: protein MID1-COMPLEMENTING ACTIVITY 2-like [Jatropha curcas] |
| 12 |
Hb_002463_050 |
0.1489125999 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_000905_100 |
0.1503911577 |
transcription factor |
TF Family: bHLH |
PREDICTED: transcription factor bHLH130-like isoform X1 [Jatropha curcas] |
| 14 |
Hb_001862_050 |
0.1509349994 |
- |
- |
PREDICTED: lysine-rich arabinogalactan protein 18-like [Jatropha curcas] |
| 15 |
Hb_004046_060 |
0.1552646088 |
- |
- |
PREDICTED: protein CDI [Jatropha curcas] |
| 16 |
Hb_001627_010 |
0.1558335534 |
- |
- |
Alpha-1,4-glucan-protein synthase [UDP-forming], putative [Ricinus communis] |
| 17 |
Hb_004223_080 |
0.1558566086 |
transcription factor |
TF Family: C2H2 |
nucleic acid binding protein, putative [Ricinus communis] |
| 18 |
Hb_000134_260 |
0.1563583352 |
transcription factor |
TF Family: bHLH |
DNA binding protein, putative [Ricinus communis] |
| 19 |
Hb_000020_080 |
0.1564402504 |
transcription factor |
TF Family: MYB |
hypothetical protein JCGZ_09080 [Jatropha curcas] |
| 20 |
Hb_000260_610 |
0.1603426706 |
- |
- |
PREDICTED: cell division cycle 20.1, cofactor of APC complex-like [Jatropha curcas] |