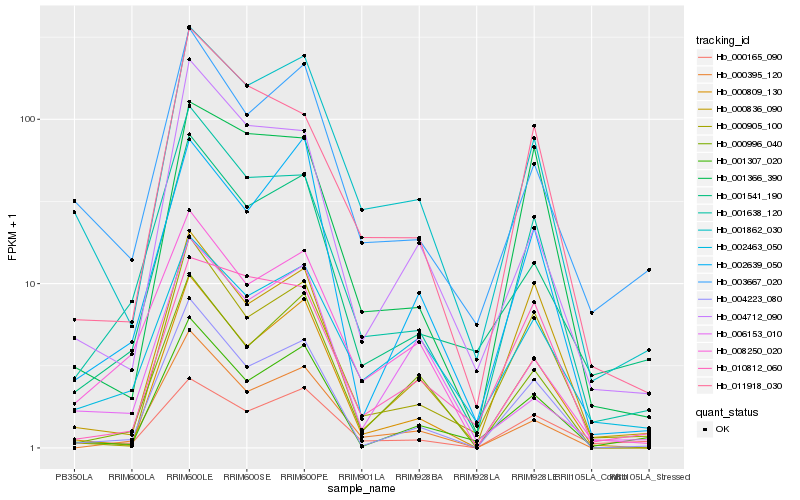
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001862_030 |
0.0 |
- |
- |
PREDICTED: probable esterase KAI2 [Jatropha curcas] |
| 2 |
Hb_003667_020 |
0.1097440624 |
- |
- |
conserved hypothetical protein 16 [Hevea brasiliensis] |
| 3 |
Hb_002463_050 |
0.133516322 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_000165_090 |
0.1387267694 |
- |
- |
PREDICTED: protein MID1-COMPLEMENTING ACTIVITY 2-like [Jatropha curcas] |
| 5 |
Hb_000809_130 |
0.1391876196 |
- |
- |
PREDICTED: peroxidase 10 isoform X3 [Jatropha curcas] |
| 6 |
Hb_001541_190 |
0.1458757068 |
- |
- |
PREDICTED: uncharacterized protein LOC105644700 [Jatropha curcas] |
| 7 |
Hb_001307_020 |
0.1503841776 |
- |
- |
electron carrier, putative [Ricinus communis] |
| 8 |
Hb_004712_090 |
0.1537089841 |
- |
- |
Serine acetyltransferase 3, mitochondrial precursor, putative [Ricinus communis] |
| 9 |
Hb_001638_120 |
0.1562738654 |
- |
- |
PREDICTED: CBL-interacting serine/threonine-protein kinase 9 [Jatropha curcas] |
| 10 |
Hb_000905_100 |
0.1590264739 |
transcription factor |
TF Family: bHLH |
PREDICTED: transcription factor bHLH130-like isoform X1 [Jatropha curcas] |
| 11 |
Hb_008250_020 |
0.159763983 |
- |
- |
PREDICTED: U-box domain-containing protein 15-like isoform X1 [Jatropha curcas] |
| 12 |
Hb_000996_040 |
0.1600293341 |
- |
- |
hypothetical protein POPTR_0003s12500g [Populus trichocarpa] |
| 13 |
Hb_004223_080 |
0.1613187327 |
transcription factor |
TF Family: C2H2 |
nucleic acid binding protein, putative [Ricinus communis] |
| 14 |
Hb_011918_030 |
0.1635301291 |
- |
- |
PREDICTED: nudix hydrolase 4 [Jatropha curcas] |
| 15 |
Hb_000395_120 |
0.1635451346 |
- |
- |
Stellacyanin, putative [Ricinus communis] |
| 16 |
Hb_002639_050 |
0.1648147793 |
- |
- |
multicopper oxidase, putative [Ricinus communis] |
| 17 |
Hb_000836_090 |
0.1711770685 |
- |
- |
PREDICTED: calmodulin-binding receptor-like cytoplasmic kinase 2 [Jatropha curcas] |
| 18 |
Hb_001366_390 |
0.1730427112 |
- |
- |
PREDICTED: uncharacterized protein LOC105642722 [Jatropha curcas] |
| 19 |
Hb_006153_010 |
0.1734911968 |
- |
- |
PREDICTED: monocopper oxidase-like protein SKU5 [Populus euphratica] |
| 20 |
Hb_010812_060 |
0.1754230184 |
- |
- |
PREDICTED: uncharacterized protein LOC105649158 [Jatropha curcas] |