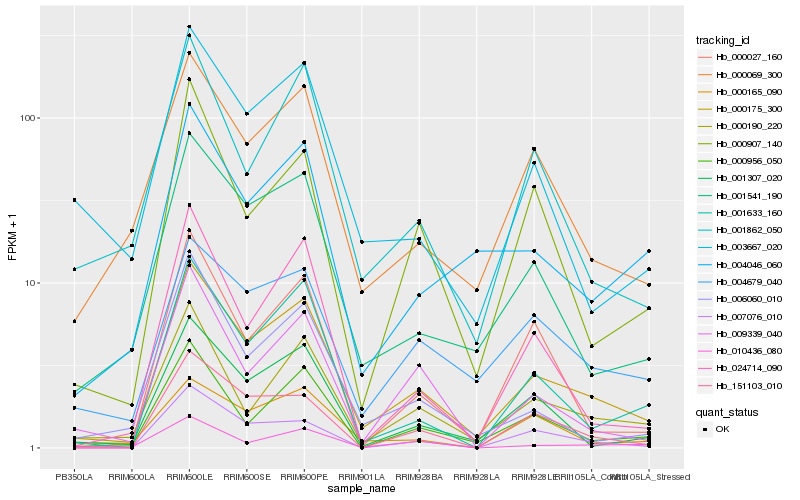
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000175_300 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105634858 [Jatropha curcas] |
| 2 |
Hb_001541_190 |
0.1267595565 |
- |
- |
PREDICTED: uncharacterized protein LOC105644700 [Jatropha curcas] |
| 3 |
Hb_000069_300 |
0.1404143748 |
- |
- |
Aspartic proteinase nepenthesin-1 precursor, putative [Ricinus communis] |
| 4 |
Hb_000190_220 |
0.1426066325 |
- |
- |
PREDICTED: serine/threonine-protein kinase Nek2 [Jatropha curcas] |
| 5 |
Hb_001307_020 |
0.1443194434 |
- |
- |
electron carrier, putative [Ricinus communis] |
| 6 |
Hb_001633_160 |
0.1477250639 |
- |
- |
PREDICTED: zerumbone synthase-like isoform X2 [Jatropha curcas] |
| 7 |
Hb_004679_040 |
0.1492227183 |
- |
- |
PREDICTED: rop guanine nucleotide exchange factor 1 [Jatropha curcas] |
| 8 |
Hb_000956_050 |
0.1505724116 |
transcription factor |
TF Family: bHLH |
PREDICTED: transcription factor SPATULA-like isoform X1 [Jatropha curcas] |
| 9 |
Hb_001862_050 |
0.1560698913 |
- |
- |
PREDICTED: lysine-rich arabinogalactan protein 18-like [Jatropha curcas] |
| 10 |
Hb_009339_040 |
0.1590094504 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_000907_140 |
0.1599182795 |
- |
- |
PREDICTED: annexin D5-like [Jatropha curcas] |
| 12 |
Hb_151103_010 |
0.1606668797 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_006060_010 |
0.1607715995 |
- |
- |
PREDICTED: cyclin-A2-2-like [Jatropha curcas] |
| 14 |
Hb_003667_020 |
0.160791727 |
- |
- |
conserved hypothetical protein 16 [Hevea brasiliensis] |
| 15 |
Hb_007076_010 |
0.1618988801 |
- |
- |
calcium-dependent protein kinase, putative [Ricinus communis] |
| 16 |
Hb_010436_080 |
0.162562825 |
- |
- |
PREDICTED: DNA replication licensing factor MCM5 [Jatropha curcas] |
| 17 |
Hb_024714_090 |
0.1638313582 |
rubber biosynthesis |
Gene Name: SRPP3 |
PREDICTED: stress-related protein [Jatropha curcas] |
| 18 |
Hb_004046_060 |
0.1674452328 |
- |
- |
PREDICTED: protein CDI [Jatropha curcas] |
| 19 |
Hb_000165_090 |
0.1688316499 |
- |
- |
PREDICTED: protein MID1-COMPLEMENTING ACTIVITY 2-like [Jatropha curcas] |
| 20 |
Hb_000027_160 |
0.1703570429 |
- |
- |
PREDICTED: probable protein phosphatase 2C 15 isoform X3 [Jatropha curcas] |