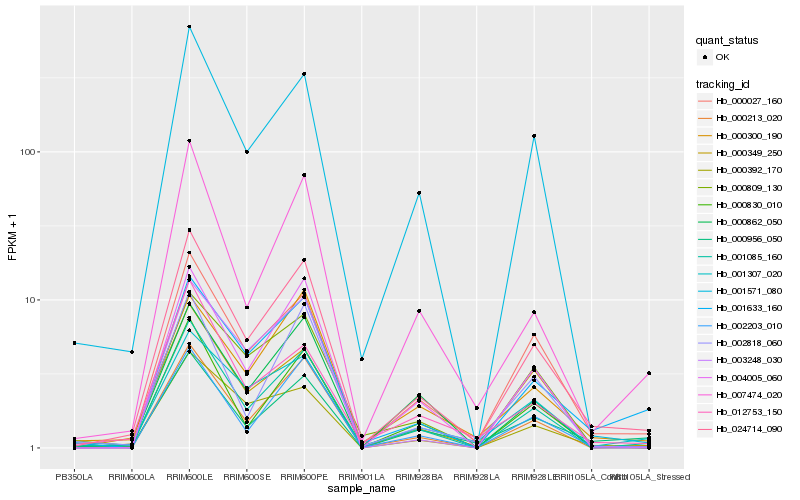
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_024714_090 |
0.0 |
rubber biosynthesis |
Gene Name: SRPP3 |
PREDICTED: stress-related protein [Jatropha curcas] |
| 2 |
Hb_001085_160 |
0.0786816109 |
- |
- |
PREDICTED: PLASMODESMATA CALLOSE-BINDING PROTEIN 3 [Jatropha curcas] |
| 3 |
Hb_000027_160 |
0.0898758176 |
- |
- |
PREDICTED: probable protein phosphatase 2C 15 isoform X3 [Jatropha curcas] |
| 4 |
Hb_000213_020 |
0.0990824094 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 5 |
Hb_001571_080 |
0.1146704647 |
- |
- |
PREDICTED: non-specific lipid-transfer protein-like protein At5g64080 [Jatropha curcas] |
| 6 |
Hb_001633_160 |
0.1159162054 |
- |
- |
PREDICTED: zerumbone synthase-like isoform X2 [Jatropha curcas] |
| 7 |
Hb_002203_010 |
0.1171803645 |
- |
- |
PREDICTED: kinesin-like protein KIF18B isoform X1 [Vitis vinifera] |
| 8 |
Hb_004005_060 |
0.1178925609 |
- |
- |
sigma factor sigb regulation protein rsbq, putative [Ricinus communis] |
| 9 |
Hb_000956_050 |
0.120959214 |
transcription factor |
TF Family: bHLH |
PREDICTED: transcription factor SPATULA-like isoform X1 [Jatropha curcas] |
| 10 |
Hb_007474_020 |
0.1212521005 |
- |
- |
multicopper oxidase, putative [Ricinus communis] |
| 11 |
Hb_000830_010 |
0.123658584 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_001307_020 |
0.1272728371 |
- |
- |
electron carrier, putative [Ricinus communis] |
| 13 |
Hb_000349_250 |
0.1300234881 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At1g12460 [Jatropha curcas] |
| 14 |
Hb_000392_170 |
0.1302105316 |
- |
- |
PREDICTED: protein PHYTOCHROME KINASE SUBSTRATE 3 [Jatropha curcas] |
| 15 |
Hb_000300_190 |
0.1302257893 |
- |
- |
PREDICTED: uncharacterized protein LOC105632566 [Jatropha curcas] |
| 16 |
Hb_003248_030 |
0.1324432937 |
- |
- |
Ent-kaurene synthase B, chloroplast precursor, putative [Ricinus communis] |
| 17 |
Hb_000862_050 |
0.1375426262 |
transcription factor |
TF Family: GRAS |
DELLA protein RGL1, putative [Ricinus communis] |
| 18 |
Hb_002818_060 |
0.1396911871 |
- |
- |
zinc finger protein, putative [Ricinus communis] |
| 19 |
Hb_000809_130 |
0.1408794049 |
- |
- |
PREDICTED: peroxidase 10 isoform X3 [Jatropha curcas] |
| 20 |
Hb_012753_150 |
0.1411989174 |
transcription factor |
TF Family: GRAS |
DELLA protein RGL1, putative [Ricinus communis] |