| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
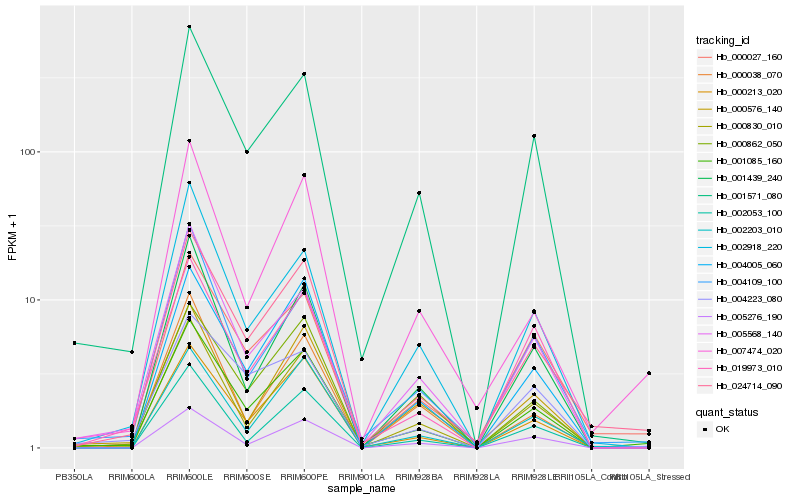
Hb_001085_160 |
0.0 |
- |
- |
PREDICTED: PLASMODESMATA CALLOSE-BINDING PROTEIN 3 [Jatropha curcas] |
| 2 |
Hb_024714_090 |
0.0786816109 |
rubber biosynthesis |
Gene Name: SRPP3 |
PREDICTED: stress-related protein [Jatropha curcas] |
| 3 |
Hb_001571_080 |
0.0817108185 |
- |
- |
PREDICTED: non-specific lipid-transfer protein-like protein At5g64080 [Jatropha curcas] |
| 4 |
Hb_000213_020 |
0.0865663162 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 5 |
Hb_000830_010 |
0.11084857 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_004005_060 |
0.1124207483 |
- |
- |
sigma factor sigb regulation protein rsbq, putative [Ricinus communis] |
| 7 |
Hb_005568_140 |
0.1151844527 |
- |
- |
ATP binding protein, putative [Ricinus communis] |
| 8 |
Hb_000027_160 |
0.1157297816 |
- |
- |
PREDICTED: probable protein phosphatase 2C 15 isoform X3 [Jatropha curcas] |
| 9 |
Hb_007474_020 |
0.1160946689 |
- |
- |
multicopper oxidase, putative [Ricinus communis] |
| 10 |
Hb_002203_010 |
0.120687941 |
- |
- |
PREDICTED: kinesin-like protein KIF18B isoform X1 [Vitis vinifera] |
| 11 |
Hb_002918_220 |
0.1214608075 |
- |
- |
PREDICTED: uncharacterized protein LOC105138896 [Populus euphratica] |
| 12 |
Hb_000862_050 |
0.1221284941 |
transcription factor |
TF Family: GRAS |
DELLA protein RGL1, putative [Ricinus communis] |
| 13 |
Hb_004109_100 |
0.123613423 |
- |
- |
Interactor of constitutive active rops 1 isoform 1 [Theobroma cacao] |
| 14 |
Hb_001439_240 |
0.1239736178 |
transcription factor |
TF Family: MYB |
hypothetical protein POPTR_0017s12230g [Populus trichocarpa] |
| 15 |
Hb_000576_140 |
0.1270012864 |
- |
- |
PREDICTED: uncharacterized protein LOC105638282 [Jatropha curcas] |
| 16 |
Hb_019973_010 |
0.1293374602 |
- |
- |
PREDICTED: probable methyltransferase PMT18 isoform X2 [Jatropha curcas] |
| 17 |
Hb_005276_190 |
0.1326794529 |
- |
- |
PREDICTED: uncharacterized protein LOC105650003 isoform X2 [Jatropha curcas] |
| 18 |
Hb_004223_080 |
0.1329422782 |
transcription factor |
TF Family: C2H2 |
nucleic acid binding protein, putative [Ricinus communis] |
| 19 |
Hb_002053_100 |
0.1350023014 |
- |
- |
actin binding protein, putative [Ricinus communis] |
| 20 |
Hb_000038_070 |
0.1364014838 |
- |
- |
conserved hypothetical protein [Ricinus communis] |