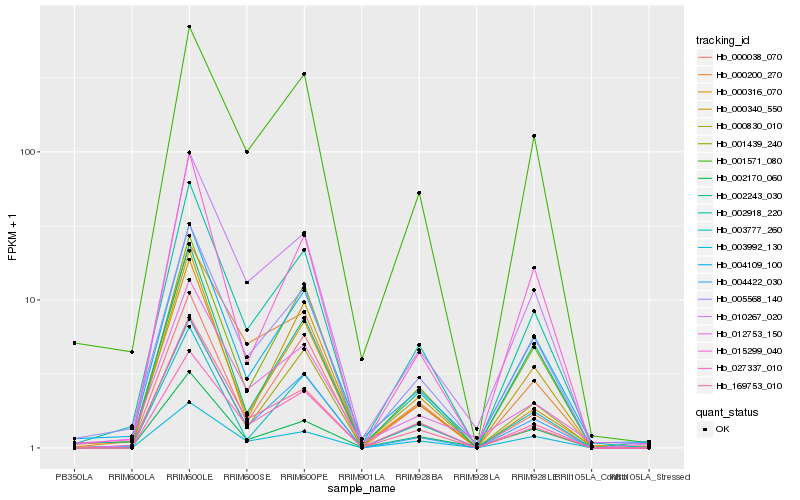
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002918_220 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105138896 [Populus euphratica] |
| 2 |
Hb_005568_140 |
0.0485392982 |
- |
- |
ATP binding protein, putative [Ricinus communis] |
| 3 |
Hb_004109_100 |
0.0724929646 |
- |
- |
Interactor of constitutive active rops 1 isoform 1 [Theobroma cacao] |
| 4 |
Hb_004422_030 |
0.0781697585 |
- |
- |
PREDICTED: serine/threonine-protein kinase SAPK7-like [Jatropha curcas] |
| 5 |
Hb_001571_080 |
0.0865008387 |
- |
- |
PREDICTED: non-specific lipid-transfer protein-like protein At5g64080 [Jatropha curcas] |
| 6 |
Hb_001439_240 |
0.0887799226 |
transcription factor |
TF Family: MYB |
hypothetical protein POPTR_0017s12230g [Populus trichocarpa] |
| 7 |
Hb_010267_020 |
0.0911018389 |
- |
- |
diphosphoinositol polyphosphate phosphohydrolase, putative [Ricinus communis] |
| 8 |
Hb_027337_010 |
0.0944130895 |
- |
- |
Potassium transporter, putative [Ricinus communis] |
| 9 |
Hb_002170_060 |
0.1014542443 |
- |
- |
PREDICTED: mitogen-activated protein kinase-binding protein 1 [Jatropha curcas] |
| 10 |
Hb_003992_130 |
0.1020531413 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 11 |
Hb_000200_270 |
0.1024075211 |
- |
- |
PREDICTED: lipid phosphate phosphatase 1-like isoform X1 [Jatropha curcas] |
| 12 |
Hb_000830_010 |
0.1032676623 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_000340_550 |
0.1041819301 |
- |
- |
PREDICTED: rho GTPase-activating protein 2 [Jatropha curcas] |
| 14 |
Hb_003777_260 |
0.1055053575 |
- |
- |
PREDICTED: uncharacterized protein LOC105640927 isoform X4 [Jatropha curcas] |
| 15 |
Hb_002243_030 |
0.1076082553 |
- |
- |
PREDICTED: 65-kDa microtubule-associated protein 3-like isoform X1 [Jatropha curcas] |
| 16 |
Hb_169753_010 |
0.109659709 |
- |
- |
hypothetical protein JCGZ_18439 [Jatropha curcas] |
| 17 |
Hb_012753_150 |
0.1113627191 |
transcription factor |
TF Family: GRAS |
DELLA protein RGL1, putative [Ricinus communis] |
| 18 |
Hb_000316_070 |
0.1117218151 |
- |
- |
PREDICTED: actin-1-like isoform X2 [Cucumis melo] |
| 19 |
Hb_015299_040 |
0.1132969118 |
transcription factor |
TF Family: MYB |
PREDICTED: myb-related protein 308 [Jatropha curcas] |
| 20 |
Hb_000038_070 |
0.1147297001 |
- |
- |
conserved hypothetical protein [Ricinus communis] |