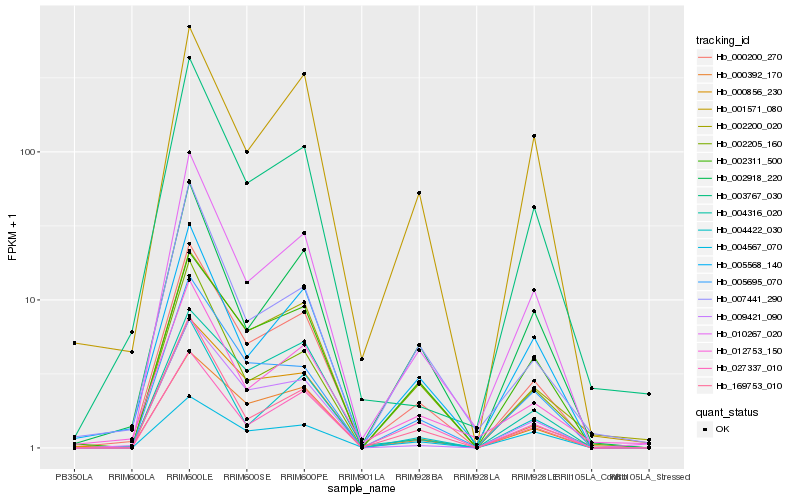
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000200_270 |
0.0 |
- |
- |
PREDICTED: lipid phosphate phosphatase 1-like isoform X1 [Jatropha curcas] |
| 2 |
Hb_010267_020 |
0.0775392186 |
- |
- |
diphosphoinositol polyphosphate phosphohydrolase, putative [Ricinus communis] |
| 3 |
Hb_000856_230 |
0.1002873285 |
- |
- |
PREDICTED: LRR receptor-like serine/threonine-protein kinase FEI 2 [Jatropha curcas] |
| 4 |
Hb_002918_220 |
0.1024075211 |
- |
- |
PREDICTED: uncharacterized protein LOC105138896 [Populus euphratica] |
| 5 |
Hb_005695_070 |
0.1050546291 |
- |
- |
Cysteine-rich RLK 29 [Theobroma cacao] |
| 6 |
Hb_002311_500 |
0.1053546008 |
- |
- |
PREDICTED: uncharacterized protein LOC105643461 [Jatropha curcas] |
| 7 |
Hb_009421_090 |
0.105453634 |
- |
- |
PREDICTED: uncharacterized protein LOC105639694 [Jatropha curcas] |
| 8 |
Hb_000392_170 |
0.107055343 |
- |
- |
PREDICTED: protein PHYTOCHROME KINASE SUBSTRATE 3 [Jatropha curcas] |
| 9 |
Hb_169753_010 |
0.1072132543 |
- |
- |
hypothetical protein JCGZ_18439 [Jatropha curcas] |
| 10 |
Hb_005568_140 |
0.1089873089 |
- |
- |
ATP binding protein, putative [Ricinus communis] |
| 11 |
Hb_002200_020 |
0.1132591185 |
transcription factor |
TF Family: C2C2-Dof |
hypothetical protein RCOM_0137680 [Ricinus communis] |
| 12 |
Hb_012753_150 |
0.1145457656 |
transcription factor |
TF Family: GRAS |
DELLA protein RGL1, putative [Ricinus communis] |
| 13 |
Hb_004422_030 |
0.1183758609 |
- |
- |
PREDICTED: serine/threonine-protein kinase SAPK7-like [Jatropha curcas] |
| 14 |
Hb_001571_080 |
0.1211448166 |
- |
- |
PREDICTED: non-specific lipid-transfer protein-like protein At5g64080 [Jatropha curcas] |
| 15 |
Hb_003767_030 |
0.1279594234 |
- |
- |
PREDICTED: probable calcium-binding protein CML23 [Jatropha curcas] |
| 16 |
Hb_007441_290 |
0.1307497383 |
- |
- |
PREDICTED: uncharacterized protein LOC105643262 [Jatropha curcas] |
| 17 |
Hb_027337_010 |
0.1312647265 |
- |
- |
Potassium transporter, putative [Ricinus communis] |
| 18 |
Hb_002205_160 |
0.1321319192 |
- |
- |
chalcone isomerase [Hibiscus cannabinus] |
| 19 |
Hb_004316_020 |
0.132464557 |
- |
- |
PREDICTED: mitogen-activated protein kinase kinase kinase NPK1-like [Jatropha curcas] |
| 20 |
Hb_004567_070 |
0.1351668836 |
- |
- |
conserved hypothetical protein [Ricinus communis] |