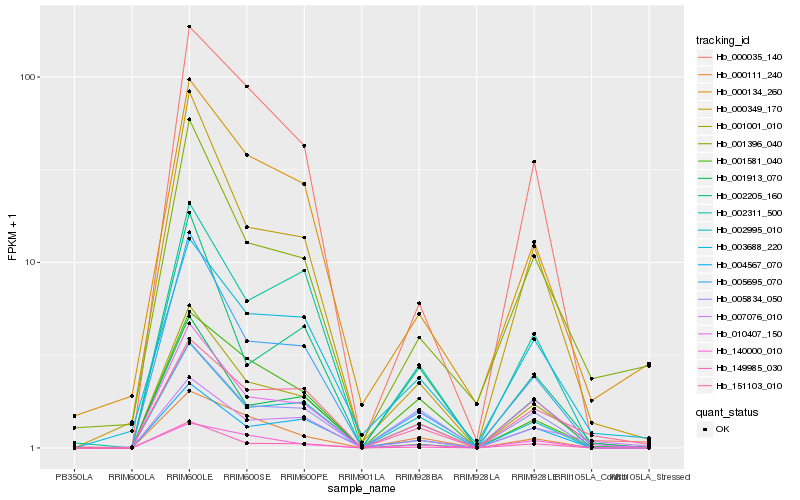
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001001_010 |
0.0 |
- |
- |
PREDICTED: patatin-like protein 2 [Jatropha curcas] |
| 2 |
Hb_010407_150 |
0.0596910712 |
transcription factor |
TF Family: MYB |
hypothetical protein POPTR_0008s16660g [Populus trichocarpa] |
| 3 |
Hb_005695_070 |
0.0864923531 |
- |
- |
Cysteine-rich RLK 29 [Theobroma cacao] |
| 4 |
Hb_149985_030 |
0.1202672713 |
- |
- |
PREDICTED: zeatin O-glucosyltransferase-like [Jatropha curcas] |
| 5 |
Hb_000349_170 |
0.1211271874 |
- |
- |
PREDICTED: geraniol 8-hydroxylase-like [Jatropha curcas] |
| 6 |
Hb_002995_010 |
0.1259220149 |
- |
- |
PREDICTED: flavonol synthase/flavanone 3-hydroxylase [Jatropha curcas] |
| 7 |
Hb_007076_010 |
0.131586124 |
- |
- |
calcium-dependent protein kinase, putative [Ricinus communis] |
| 8 |
Hb_004567_070 |
0.1330812788 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 9 |
Hb_000035_140 |
0.1355537159 |
transcription factor |
TF Family: WRKY |
putative WRKY transcription factor family protein [Populus trichocarpa] |
| 10 |
Hb_000111_240 |
0.1374851161 |
- |
- |
PREDICTED: glutamate receptor 2.8-like [Elaeis guineensis] |
| 11 |
Hb_002311_500 |
0.138086189 |
- |
- |
PREDICTED: uncharacterized protein LOC105643461 [Jatropha curcas] |
| 12 |
Hb_151103_010 |
0.1409530022 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_001396_040 |
0.1419403395 |
- |
- |
PREDICTED: uncharacterized protein LOC105644845 [Jatropha curcas] |
| 14 |
Hb_002205_160 |
0.1428896956 |
- |
- |
chalcone isomerase [Hibiscus cannabinus] |
| 15 |
Hb_003688_220 |
0.1435562645 |
- |
- |
unknown [Populus trichocarpa] |
| 16 |
Hb_005834_050 |
0.1444975918 |
- |
- |
PREDICTED: cationic amino acid transporter 1-like [Jatropha curcas] |
| 17 |
Hb_000134_260 |
0.1471907806 |
transcription factor |
TF Family: bHLH |
DNA binding protein, putative [Ricinus communis] |
| 18 |
Hb_140000_010 |
0.1476286926 |
- |
- |
receptor-kinase, putative [Ricinus communis] |
| 19 |
Hb_001913_070 |
0.1485913665 |
- |
- |
PREDICTED: caffeoylshikimate esterase-like [Jatropha curcas] |
| 20 |
Hb_001581_040 |
0.1489028244 |
- |
- |
BRASSINOSTEROID INSENSITIVE 1-associated receptor kinase 1 precursor, putative [Ricinus communis] |