| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
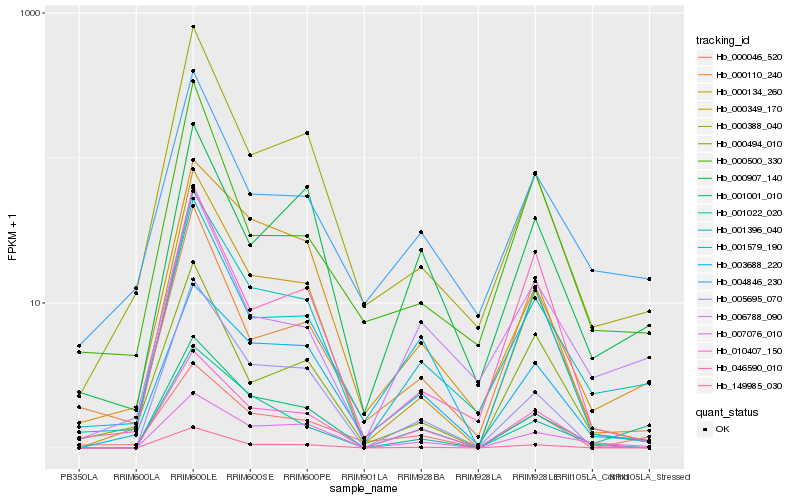
Hb_001396_040 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105644845 [Jatropha curcas] |
| 2 |
Hb_004846_230 |
0.121275935 |
- |
- |
Chaperone protein dnaJ 8, chloroplast precursor, putative [Ricinus communis] |
| 3 |
Hb_000388_040 |
0.1322258401 |
- |
- |
PREDICTED: probable calcium-binding protein CML23 [Jatropha curcas] |
| 4 |
Hb_006788_090 |
0.1350997163 |
- |
- |
- |
| 5 |
Hb_000134_260 |
0.1379410179 |
transcription factor |
TF Family: bHLH |
DNA binding protein, putative [Ricinus communis] |
| 6 |
Hb_001001_010 |
0.1419403395 |
- |
- |
PREDICTED: patatin-like protein 2 [Jatropha curcas] |
| 7 |
Hb_000349_170 |
0.1436980846 |
- |
- |
PREDICTED: geraniol 8-hydroxylase-like [Jatropha curcas] |
| 8 |
Hb_007076_010 |
0.147190007 |
- |
- |
calcium-dependent protein kinase, putative [Ricinus communis] |
| 9 |
Hb_010407_150 |
0.1474300416 |
transcription factor |
TF Family: MYB |
hypothetical protein POPTR_0008s16660g [Populus trichocarpa] |
| 10 |
Hb_000907_140 |
0.1529202444 |
- |
- |
PREDICTED: annexin D5-like [Jatropha curcas] |
| 11 |
Hb_001579_190 |
0.1542451852 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_005695_070 |
0.1545934364 |
- |
- |
Cysteine-rich RLK 29 [Theobroma cacao] |
| 13 |
Hb_003688_220 |
0.1549230746 |
- |
- |
unknown [Populus trichocarpa] |
| 14 |
Hb_000494_010 |
0.1622495812 |
transcription factor |
TF Family: HB |
hypothetical protein POPTR_0014s04460g [Populus trichocarpa] |
| 15 |
Hb_000500_330 |
0.1629617895 |
- |
- |
PREDICTED: bidirectional sugar transporter SWEET2a-like [Jatropha curcas] |
| 16 |
Hb_000046_520 |
0.1638831174 |
- |
- |
PREDICTED: proline-rich receptor-like protein kinase PERK3 isoform X1 [Jatropha curcas] |
| 17 |
Hb_149985_030 |
0.1665037994 |
- |
- |
PREDICTED: zeatin O-glucosyltransferase-like [Jatropha curcas] |
| 18 |
Hb_001022_020 |
0.1665373389 |
transcription factor |
TF Family: NAC |
NAC transcription factor 038 [Jatropha curcas] |
| 19 |
Hb_000110_240 |
0.1674099598 |
- |
- |
PREDICTED: uncharacterized protein LOC105642006 isoform X3 [Jatropha curcas] |
| 20 |
Hb_046590_010 |
0.169730507 |
- |
- |
Cycloeucalenol cycloisomerase, putative [Ricinus communis] |