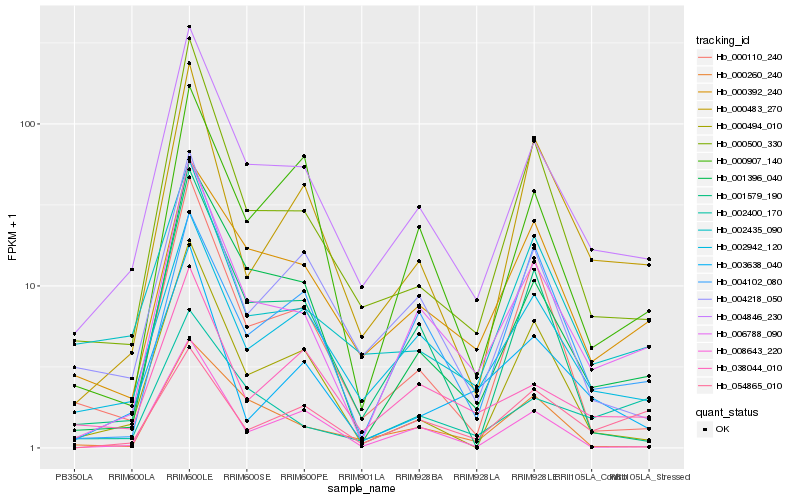
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_006788_090 |
0.0 |
- |
- |
- |
| 2 |
Hb_004846_230 |
0.1114241465 |
- |
- |
Chaperone protein dnaJ 8, chloroplast precursor, putative [Ricinus communis] |
| 3 |
Hb_001396_040 |
0.1350997163 |
- |
- |
PREDICTED: uncharacterized protein LOC105644845 [Jatropha curcas] |
| 4 |
Hb_000500_330 |
0.1570109427 |
- |
- |
PREDICTED: bidirectional sugar transporter SWEET2a-like [Jatropha curcas] |
| 5 |
Hb_002942_120 |
0.158080596 |
transcription factor |
TF Family: bHLH |
Transcription factor ICE1, putative [Ricinus communis] |
| 6 |
Hb_000483_270 |
0.1732665573 |
- |
- |
PREDICTED: uncharacterized protein LOC105632352 [Jatropha curcas] |
| 7 |
Hb_000392_240 |
0.1786373182 |
- |
- |
DNA binding protein, putative [Ricinus communis] |
| 8 |
Hb_001579_190 |
0.1788907424 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 9 |
Hb_002400_170 |
0.1794662203 |
- |
- |
PREDICTED: adenine phosphoribosyltransferase 5 [Jatropha curcas] |
| 10 |
Hb_008643_220 |
0.1842530423 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At1g53440 [Jatropha curcas] |
| 11 |
Hb_000110_240 |
0.185125437 |
- |
- |
PREDICTED: uncharacterized protein LOC105642006 isoform X3 [Jatropha curcas] |
| 12 |
Hb_038044_010 |
0.1890722098 |
- |
- |
12-oxophytodienoate reductase 2 [Theobroma cacao] |
| 13 |
Hb_000907_140 |
0.1901388 |
- |
- |
PREDICTED: annexin D5-like [Jatropha curcas] |
| 14 |
Hb_000260_240 |
0.1937654892 |
- |
- |
hypothetical protein JCGZ_00622 [Jatropha curcas] |
| 15 |
Hb_003638_040 |
0.1968854682 |
- |
- |
kinesin heavy chain, putative [Ricinus communis] |
| 16 |
Hb_000494_010 |
0.1982964947 |
transcription factor |
TF Family: HB |
hypothetical protein POPTR_0014s04460g [Populus trichocarpa] |
| 17 |
Hb_054865_010 |
0.1989753632 |
- |
- |
PREDICTED: uncharacterized membrane protein At1g16860-like [Jatropha curcas] |
| 18 |
Hb_004218_050 |
0.199973794 |
- |
- |
annexin [Manihot esculenta] |
| 19 |
Hb_004102_080 |
0.2013563842 |
- |
- |
PREDICTED: acyl-protein thioesterase 2 [Jatropha curcas] |
| 20 |
Hb_002435_090 |
0.2023400179 |
- |
- |
PREDICTED: membrane-associated 30 kDa protein, chloroplastic [Jatropha curcas] |