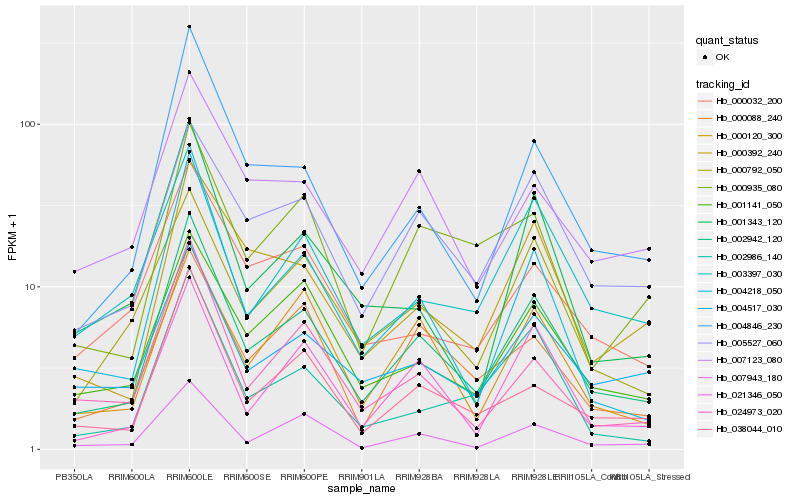
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002942_120 |
0.0 |
transcription factor |
TF Family: bHLH |
Transcription factor ICE1, putative [Ricinus communis] |
| 2 |
Hb_004218_050 |
0.1055875261 |
- |
- |
annexin [Manihot esculenta] |
| 3 |
Hb_004846_230 |
0.1153429355 |
- |
- |
Chaperone protein dnaJ 8, chloroplast precursor, putative [Ricinus communis] |
| 4 |
Hb_038044_010 |
0.121347851 |
- |
- |
12-oxophytodienoate reductase 2 [Theobroma cacao] |
| 5 |
Hb_005527_060 |
0.1272122035 |
- |
- |
malic enzyme, putative [Ricinus communis] |
| 6 |
Hb_021346_050 |
0.1287003383 |
- |
- |
xylulose kinase, putative [Ricinus communis] |
| 7 |
Hb_007943_180 |
0.1290898457 |
- |
- |
PREDICTED: uncharacterized protein LOC105637734 [Jatropha curcas] |
| 8 |
Hb_001141_050 |
0.1330832273 |
transcription factor |
TF Family: CPP |
PREDICTED: protein tesmin/TSO1-like CXC 2 isoform X2 [Jatropha curcas] |
| 9 |
Hb_000792_050 |
0.1330873278 |
- |
- |
PREDICTED: acetyl-coenzyme A carboxylase carboxyl transferase subunit alpha, chloroplastic isoform X1 [Jatropha curcas] |
| 10 |
Hb_000392_240 |
0.1362974721 |
- |
- |
DNA binding protein, putative [Ricinus communis] |
| 11 |
Hb_004517_030 |
0.136690319 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_000120_300 |
0.1379927095 |
- |
- |
PREDICTED: kinesin-like calmodulin-binding protein isoform X1 [Eucalyptus grandis] |
| 13 |
Hb_024973_020 |
0.1383938852 |
- |
- |
Protein regulator of cytokinesis, putative [Ricinus communis] |
| 14 |
Hb_007123_080 |
0.1404898137 |
- |
- |
big map kinase/bmk, putative [Ricinus communis] |
| 15 |
Hb_000088_240 |
0.1411150888 |
transcription factor |
TF Family: MYB |
r2r3-myb transcription factor, putative [Ricinus communis] |
| 16 |
Hb_003397_030 |
0.1417468648 |
rubber biosynthesis |
Gene Name: Pyruvate dehydrogenase |
PREDICTED: pyruvate dehydrogenase E1 component subunit beta-3, chloroplastic [Jatropha curcas] |
| 17 |
Hb_002986_140 |
0.1422967566 |
- |
- |
PREDICTED: uncharacterized protein LOC105639684 [Jatropha curcas] |
| 18 |
Hb_001343_120 |
0.142459344 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 19 |
Hb_000935_080 |
0.1442225615 |
- |
- |
PREDICTED: branched-chain-amino-acid aminotransferase 3, chloroplastic isoform X1 [Jatropha curcas] |
| 20 |
Hb_000032_200 |
0.1469230084 |
- |
- |
PREDICTED: probable polygalacturonase [Jatropha curcas] |