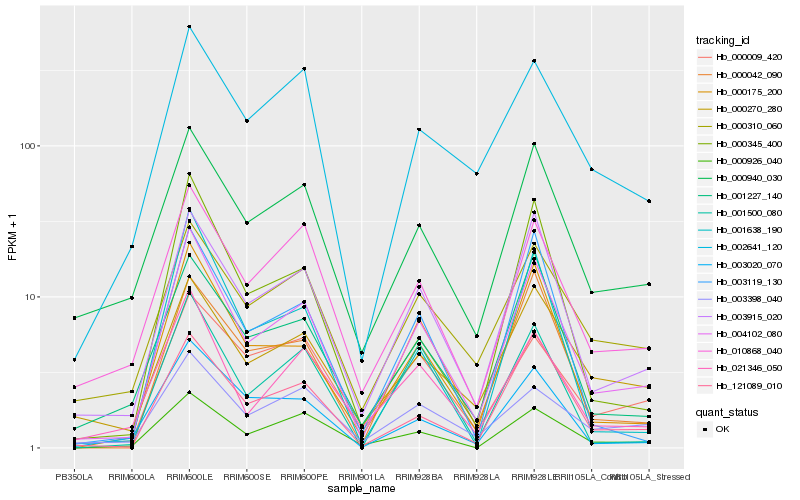
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_004102_080 |
0.0 |
- |
- |
PREDICTED: acyl-protein thioesterase 2 [Jatropha curcas] |
| 2 |
Hb_003915_020 |
0.0962988121 |
- |
- |
PREDICTED: outer envelope pore protein 37, chloroplastic [Jatropha curcas] |
| 3 |
Hb_000175_200 |
0.1146292831 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_121089_010 |
0.1203102439 |
- |
- |
PREDICTED: uncharacterized protein LOC105645663 [Jatropha curcas] |
| 5 |
Hb_000926_040 |
0.1289725362 |
- |
- |
PREDICTED: DNA mismatch repair protein MSH5 [Jatropha curcas] |
| 6 |
Hb_021346_050 |
0.1291851156 |
- |
- |
xylulose kinase, putative [Ricinus communis] |
| 7 |
Hb_010868_040 |
0.1332045128 |
- |
- |
PREDICTED: reticulon-like protein B21 isoform X1 [Populus euphratica] |
| 8 |
Hb_000940_030 |
0.1336224672 |
- |
- |
PREDICTED: triosephosphate isomerase, chloroplastic [Jatropha curcas] |
| 9 |
Hb_001227_140 |
0.1370119761 |
- |
- |
PREDICTED: protein ROS1 [Jatropha curcas] |
| 10 |
Hb_003020_070 |
0.1391170779 |
- |
- |
PREDICTED: uncharacterized protein LOC103997323 [Musa acuminata subsp. malaccensis] |
| 11 |
Hb_003119_130 |
0.1426218523 |
- |
- |
hypothetical protein POPTR_0017s07770g [Populus trichocarpa] |
| 12 |
Hb_000270_280 |
0.1430906022 |
- |
- |
Rhicadhesin receptor precursor, putative [Ricinus communis] |
| 13 |
Hb_000345_400 |
0.1463358954 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 14 |
Hb_000009_420 |
0.1475654947 |
- |
- |
PREDICTED: uncharacterized protein LOC105639614 isoform X2 [Jatropha curcas] |
| 15 |
Hb_001500_080 |
0.147887997 |
- |
- |
PREDICTED: uncharacterized protein LOC105647337 isoform X1 [Jatropha curcas] |
| 16 |
Hb_002641_120 |
0.1485564245 |
- |
- |
PREDICTED: calmodulin [Jatropha curcas] |
| 17 |
Hb_001638_190 |
0.1492547667 |
- |
- |
PREDICTED: protein IQ-DOMAIN 1-like isoform X1 [Jatropha curcas] |
| 18 |
Hb_000042_090 |
0.1493119435 |
desease resistance |
Gene Name: NB-ARC |
Disease resistance protein RPS2, putative [Ricinus communis] |
| 19 |
Hb_000310_060 |
0.1493789648 |
- |
- |
hypothetical protein JCGZ_20793 [Jatropha curcas] |
| 20 |
Hb_003398_040 |
0.1507509941 |
- |
- |
PREDICTED: actin-related protein 5 isoform X1 [Jatropha curcas] |