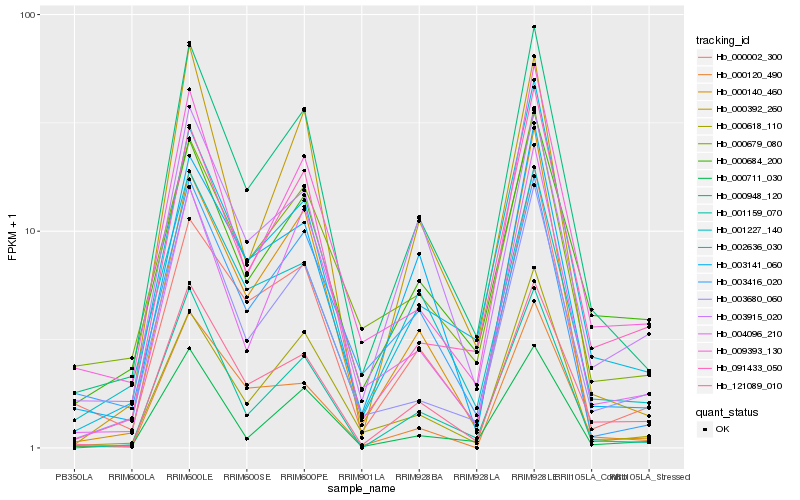
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000948_120 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105634688 isoform X1 [Jatropha curcas] |
| 2 |
Hb_002636_030 |
0.0932092152 |
- |
- |
hypothetical protein JCGZ_11800 [Jatropha curcas] |
| 3 |
Hb_121089_010 |
0.0948292051 |
- |
- |
PREDICTED: uncharacterized protein LOC105645663 [Jatropha curcas] |
| 4 |
Hb_001227_140 |
0.103955852 |
- |
- |
PREDICTED: protein ROS1 [Jatropha curcas] |
| 5 |
Hb_001159_070 |
0.104055508 |
- |
- |
DNA photolyase, putative [Ricinus communis] |
| 6 |
Hb_003680_060 |
0.1045087778 |
- |
- |
hypothetical protein VITISV_022322 [Vitis vinifera] |
| 7 |
Hb_000140_460 |
0.1056820739 |
- |
- |
PREDICTED: uncharacterized protein LOC105628417 [Jatropha curcas] |
| 8 |
Hb_003141_060 |
0.1060071889 |
transcription factor |
TF Family: TRAF |
PREDICTED: ETO1-like protein 1 [Jatropha curcas] |
| 9 |
Hb_009393_130 |
0.1072664026 |
- |
- |
PREDICTED: uncharacterized protein LOC105638053 isoform X4 [Jatropha curcas] |
| 10 |
Hb_000618_110 |
0.1103555785 |
- |
- |
hypothetical protein POPTR_0003s09870g [Populus trichocarpa] |
| 11 |
Hb_000002_300 |
0.1108707281 |
- |
- |
Desacetoxyvindoline 4-hydroxylase, putative [Ricinus communis] |
| 12 |
Hb_091433_050 |
0.1128340494 |
- |
- |
PREDICTED: uncharacterized protein LOC105646626 [Jatropha curcas] |
| 13 |
Hb_003915_020 |
0.1158636591 |
- |
- |
PREDICTED: outer envelope pore protein 37, chloroplastic [Jatropha curcas] |
| 14 |
Hb_000392_260 |
0.116107373 |
- |
- |
PREDICTED: microtubule-associated protein RP/EB family member 1B [Jatropha curcas] |
| 15 |
Hb_000679_080 |
0.1174506947 |
- |
- |
PREDICTED: lactation elevated protein 1 isoform X1 [Jatropha curcas] |
| 16 |
Hb_003416_020 |
0.1192135717 |
- |
- |
pentatricopeptide repeat-containing family protein [Populus trichocarpa] |
| 17 |
Hb_000711_030 |
0.1200264288 |
- |
- |
hypothetical protein POPTR_0019s02860g [Populus trichocarpa] |
| 18 |
Hb_004096_210 |
0.1205845693 |
- |
- |
PREDICTED: uncharacterized protein LOC105638707 isoform X2 [Jatropha curcas] |
| 19 |
Hb_000684_200 |
0.1230897145 |
- |
- |
PREDICTED: protein IQ-DOMAIN 1-like [Jatropha curcas] |
| 20 |
Hb_000120_490 |
0.1234611022 |
- |
- |
PREDICTED: MORC family CW-type zinc finger protein 3-like [Jatropha curcas] |