| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
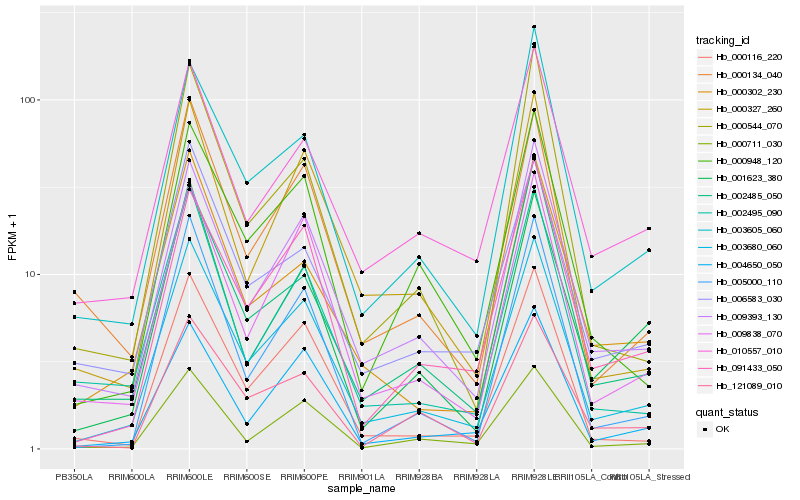
Hb_003680_060 |
0.0 |
- |
- |
hypothetical protein VITISV_022322 [Vitis vinifera] |
| 2 |
Hb_009393_130 |
0.0775841284 |
- |
- |
PREDICTED: uncharacterized protein LOC105638053 isoform X4 [Jatropha curcas] |
| 3 |
Hb_009838_070 |
0.0848151813 |
- |
- |
PREDICTED: 28 kDa ribonucleoprotein, chloroplastic [Jatropha curcas] |
| 4 |
Hb_000116_220 |
0.0937326775 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 5 |
Hb_004650_050 |
0.0956501713 |
- |
- |
PREDICTED: uncharacterized protein At1g04910-like isoform X2 [Jatropha curcas] |
| 6 |
Hb_091433_050 |
0.1004166704 |
- |
- |
PREDICTED: uncharacterized protein LOC105646626 [Jatropha curcas] |
| 7 |
Hb_000711_030 |
0.101289934 |
- |
- |
hypothetical protein POPTR_0019s02860g [Populus trichocarpa] |
| 8 |
Hb_000327_260 |
0.1025026552 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 9 |
Hb_003605_060 |
0.1028641207 |
- |
- |
PREDICTED: ATP synthase subunit delta, chloroplastic [Jatropha curcas] |
| 10 |
Hb_000948_120 |
0.1045087778 |
- |
- |
PREDICTED: uncharacterized protein LOC105634688 isoform X1 [Jatropha curcas] |
| 11 |
Hb_000134_040 |
0.1063128375 |
- |
- |
PREDICTED: 30S ribosomal protein 2, chloroplastic [Jatropha curcas] |
| 12 |
Hb_005000_110 |
0.1098594978 |
- |
- |
PREDICTED: uncharacterized protein LOC105637872 isoform X1 [Jatropha curcas] |
| 13 |
Hb_002485_050 |
0.1107099853 |
- |
- |
PREDICTED: probable plastid-lipid-associated protein 12, chloroplastic [Jatropha curcas] |
| 14 |
Hb_002495_090 |
0.110809014 |
transcription factor |
TF Family: bHLH |
PREDICTED: transcription factor bHLH48 isoform X2 [Jatropha curcas] |
| 15 |
Hb_006583_030 |
0.1108307517 |
- |
- |
PREDICTED: SPX and EXS domain-containing protein 1-like isoform X4 [Jatropha curcas] |
| 16 |
Hb_000302_230 |
0.1114033514 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_010557_010 |
0.1171672122 |
- |
- |
PREDICTED: 50S ribosomal protein L3, chloroplastic [Jatropha curcas] |
| 18 |
Hb_001623_380 |
0.1176566198 |
- |
- |
PREDICTED: 7-hydroxymethyl chlorophyll a reductase, chloroplastic [Jatropha curcas] |
| 19 |
Hb_121089_010 |
0.1179845386 |
- |
- |
PREDICTED: uncharacterized protein LOC105645663 [Jatropha curcas] |
| 20 |
Hb_000544_070 |
0.1200988402 |
- |
- |
Peptidyl-prolyl cis-trans isomerase FKBP17-2 [Morus notabilis] |