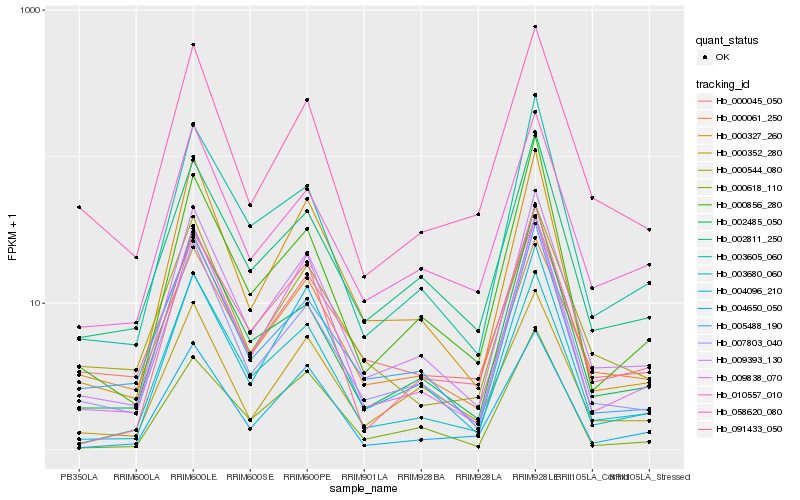
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_009393_130 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105638053 isoform X4 [Jatropha curcas] |
| 2 |
Hb_003680_060 |
0.0775841284 |
- |
- |
hypothetical protein VITISV_022322 [Vitis vinifera] |
| 3 |
Hb_007803_040 |
0.0792390296 |
- |
- |
PREDICTED: peptidyl-prolyl cis-trans isomerase CYP37, chloroplastic [Jatropha curcas] |
| 4 |
Hb_003605_060 |
0.0807991779 |
- |
- |
PREDICTED: ATP synthase subunit delta, chloroplastic [Jatropha curcas] |
| 5 |
Hb_009838_070 |
0.0816972979 |
- |
- |
PREDICTED: 28 kDa ribonucleoprotein, chloroplastic [Jatropha curcas] |
| 6 |
Hb_010557_010 |
0.085319044 |
- |
- |
PREDICTED: 50S ribosomal protein L3, chloroplastic [Jatropha curcas] |
| 7 |
Hb_002811_250 |
0.0863750436 |
- |
- |
PREDICTED: probable ribose-5-phosphate isomerase 3, chloroplastic [Jatropha curcas] |
| 8 |
Hb_002485_050 |
0.089597695 |
- |
- |
PREDICTED: probable plastid-lipid-associated protein 12, chloroplastic [Jatropha curcas] |
| 9 |
Hb_000327_260 |
0.0922424297 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 10 |
Hb_058620_080 |
0.0940422548 |
- |
- |
PREDICTED: phosphoglycerate kinase, chloroplastic [Jatropha curcas] |
| 11 |
Hb_004096_210 |
0.0941332696 |
- |
- |
PREDICTED: uncharacterized protein LOC105638707 isoform X2 [Jatropha curcas] |
| 12 |
Hb_005488_190 |
0.0956685356 |
- |
- |
RNA polymerase sigma factor rpoD, putative [Ricinus communis] |
| 13 |
Hb_000352_280 |
0.0991540476 |
- |
- |
PREDICTED: acetyl-coenzyme A synthetase, chloroplastic/glyoxysomal isoform X2 [Jatropha curcas] |
| 14 |
Hb_000544_080 |
0.1011072257 |
- |
- |
PREDICTED: uncharacterized protein LOC105631062 isoform X3 [Jatropha curcas] |
| 15 |
Hb_091433_050 |
0.1011381546 |
- |
- |
PREDICTED: uncharacterized protein LOC105646626 [Jatropha curcas] |
| 16 |
Hb_000856_280 |
0.1017522603 |
- |
- |
PREDICTED: uncharacterized protein LOC105640491 [Jatropha curcas] |
| 17 |
Hb_004650_050 |
0.1055500986 |
- |
- |
PREDICTED: uncharacterized protein At1g04910-like isoform X2 [Jatropha curcas] |
| 18 |
Hb_000618_110 |
0.1064381891 |
- |
- |
hypothetical protein POPTR_0003s09870g [Populus trichocarpa] |
| 19 |
Hb_000045_050 |
0.1069692049 |
- |
- |
PREDICTED: probable zinc metalloprotease EGY2, chloroplastic [Jatropha curcas] |
| 20 |
Hb_000061_250 |
0.1070716851 |
- |
- |
hypothetical protein JCGZ_11665 [Jatropha curcas] |