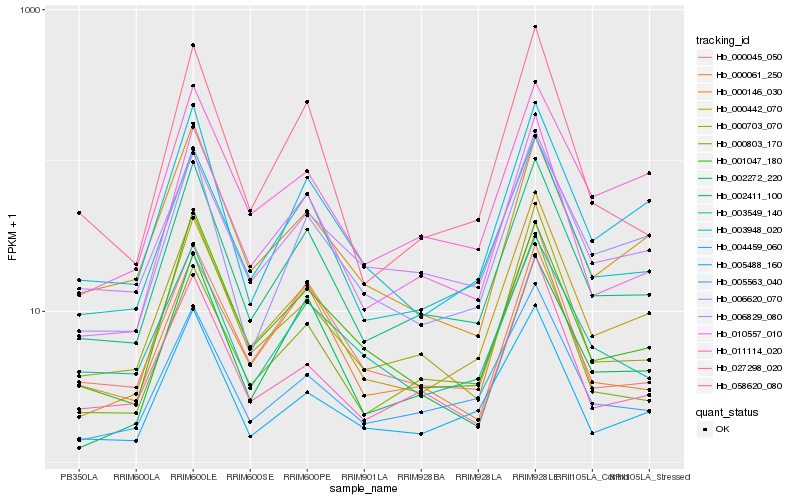
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002411_100 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105631266 [Jatropha curcas] |
| 2 |
Hb_003948_020 |
0.0621574458 |
- |
- |
PREDICTED: transmembrane protein 41B [Jatropha curcas] |
| 3 |
Hb_010557_010 |
0.0682247229 |
- |
- |
PREDICTED: 50S ribosomal protein L3, chloroplastic [Jatropha curcas] |
| 4 |
Hb_000703_070 |
0.083249723 |
- |
- |
PREDICTED: uncharacterized protein LOC105635704 isoform X1 [Jatropha curcas] |
| 5 |
Hb_001047_180 |
0.0890386518 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_004459_060 |
0.0937877748 |
- |
- |
50S ribosomal protein L34, chloroplast precursor, putative [Ricinus communis] |
| 7 |
Hb_005563_040 |
0.0957896344 |
- |
- |
PREDICTED: uncharacterized protein LOC105630105 isoform X1 [Jatropha curcas] |
| 8 |
Hb_003549_140 |
0.100185796 |
- |
- |
Thylakoid lumenal 21.5 kDa protein, chloroplast precursor, putative [Ricinus communis] |
| 9 |
Hb_011114_020 |
0.1009682123 |
- |
- |
PREDICTED: 50S ribosomal protein L12-1, chloroplastic-like [Jatropha curcas] |
| 10 |
Hb_000045_050 |
0.1014124548 |
- |
- |
PREDICTED: probable zinc metalloprotease EGY2, chloroplastic [Jatropha curcas] |
| 11 |
Hb_005488_160 |
0.1020736786 |
- |
- |
PREDICTED: maltose excess protein 1, chloroplastic-like [Jatropha curcas] |
| 12 |
Hb_000442_070 |
0.1026993513 |
- |
- |
PREDICTED: uncharacterized protein LOC105645538 [Jatropha curcas] |
| 13 |
Hb_006620_070 |
0.1039158969 |
- |
- |
chloroplast 30S ribosomal protein S13 [Hevea brasiliensis] |
| 14 |
Hb_002272_220 |
0.1047394021 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 15 |
Hb_000146_030 |
0.1048697709 |
- |
- |
PREDICTED: 50S ribosomal protein L10, chloroplastic [Jatropha curcas] |
| 16 |
Hb_000803_170 |
0.1051063222 |
- |
- |
PREDICTED: uncharacterized protein LOC105648312 [Jatropha curcas] |
| 17 |
Hb_000061_250 |
0.1051300094 |
- |
- |
hypothetical protein JCGZ_11665 [Jatropha curcas] |
| 18 |
Hb_058620_080 |
0.105543262 |
- |
- |
PREDICTED: phosphoglycerate kinase, chloroplastic [Jatropha curcas] |
| 19 |
Hb_006829_080 |
0.1064947893 |
- |
- |
PREDICTED: protein LHCP TRANSLOCATION DEFECT [Jatropha curcas] |
| 20 |
Hb_027298_020 |
0.1101101268 |
- |
- |
hypothetical protein POPTR_0005s05760g [Populus trichocarpa] |