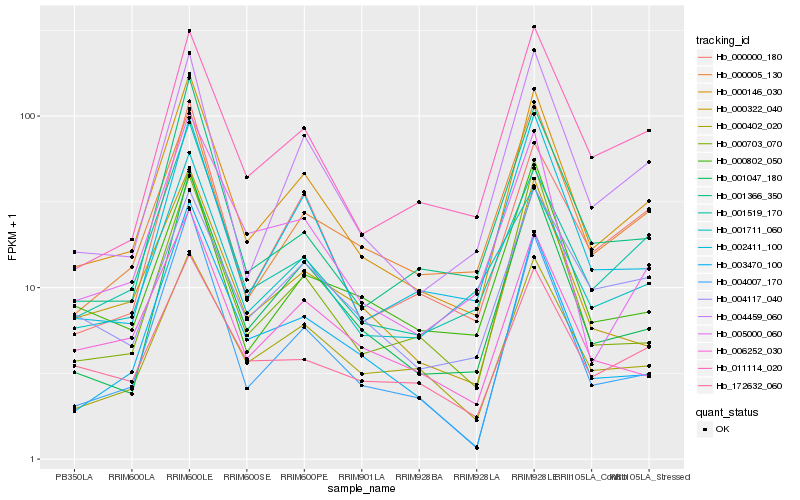
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000146_030 |
0.0 |
- |
- |
PREDICTED: 50S ribosomal protein L10, chloroplastic [Jatropha curcas] |
| 2 |
Hb_001047_180 |
0.0877505686 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_004459_060 |
0.0933612136 |
- |
- |
50S ribosomal protein L34, chloroplast precursor, putative [Ricinus communis] |
| 4 |
Hb_002411_100 |
0.1048697709 |
- |
- |
PREDICTED: uncharacterized protein LOC105631266 [Jatropha curcas] |
| 5 |
Hb_000402_020 |
0.1052904624 |
- |
- |
PREDICTED: 2-phytyl-1,4-beta-naphthoquinone methyltransferase, chloroplastic isoform X1 [Jatropha curcas] |
| 6 |
Hb_004007_170 |
0.1057276991 |
- |
- |
Glucan endo-1,3-beta-glucosidase precursor, putative [Ricinus communis] |
| 7 |
Hb_000322_040 |
0.1072835185 |
- |
- |
PREDICTED: lycopene epsilon cyclase, chloroplastic [Jatropha curcas] |
| 8 |
Hb_000005_130 |
0.1094891569 |
- |
- |
hypothetical protein POPTR_0004s22660g [Populus trichocarpa] |
| 9 |
Hb_011114_020 |
0.109899216 |
- |
- |
PREDICTED: 50S ribosomal protein L12-1, chloroplastic-like [Jatropha curcas] |
| 10 |
Hb_001519_170 |
0.1099367685 |
- |
- |
PREDICTED: K(+) efflux antiporter 3, chloroplastic [Jatropha curcas] |
| 11 |
Hb_006252_030 |
0.1116787488 |
- |
- |
PREDICTED: uncharacterized protein LOC105638333 [Jatropha curcas] |
| 12 |
Hb_000000_180 |
0.1118234924 |
- |
- |
PREDICTED: 30S ribosomal protein S9, chloroplastic-like [Populus euphratica] |
| 13 |
Hb_000703_070 |
0.111855761 |
- |
- |
PREDICTED: uncharacterized protein LOC105635704 isoform X1 [Jatropha curcas] |
| 14 |
Hb_005000_060 |
0.114922631 |
- |
- |
PREDICTED: uncharacterized protein LOC105637867 [Jatropha curcas] |
| 15 |
Hb_004117_040 |
0.1179633204 |
- |
- |
PREDICTED: uncharacterized protein LOC105638287 [Jatropha curcas] |
| 16 |
Hb_003470_100 |
0.1182819509 |
- |
- |
protein binding protein, putative [Ricinus communis] |
| 17 |
Hb_001711_060 |
0.1190232843 |
- |
- |
PREDICTED: ferredoxin isoform X1 [Jatropha curcas] |
| 18 |
Hb_001366_350 |
0.1197082748 |
- |
- |
PREDICTED: peptidyl-prolyl cis-trans isomerase isoform X2 [Jatropha curcas] |
| 19 |
Hb_172632_060 |
0.1229723716 |
- |
- |
PREDICTED: uncharacterized protein LOC105646135 isoform X1 [Jatropha curcas] |
| 20 |
Hb_000802_050 |
0.1236775364 |
transcription factor |
TF Family: TCP |
PREDICTED: transcription factor TCP2-like [Jatropha curcas] |