| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
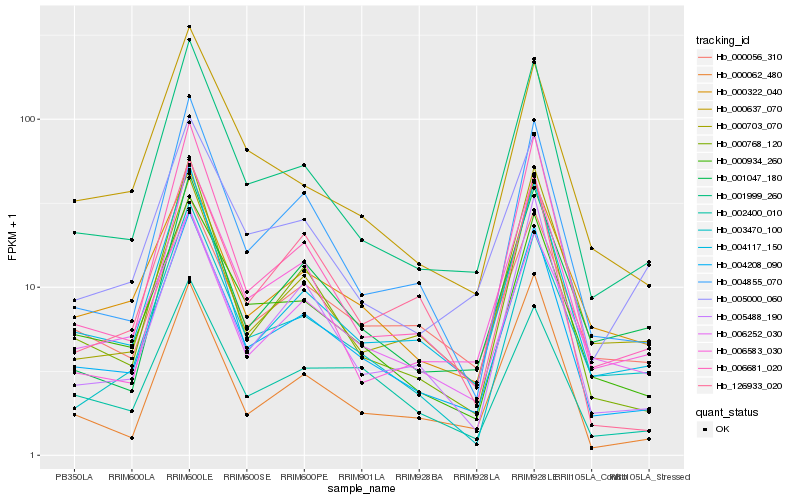
Hb_004208_090 |
0.0 |
- |
- |
RNA polymerase sigma factor rpoD, putative [Ricinus communis] |
| 2 |
Hb_001999_260 |
0.0565119339 |
- |
- |
PREDICTED: 50S ribosomal protein L4, chloroplastic [Jatropha curcas] |
| 3 |
Hb_000934_260 |
0.077198878 |
- |
- |
Thylakoid lumenal 29.8 kDa protein, chloroplast precursor, putative [Ricinus communis] |
| 4 |
Hb_004117_150 |
0.0791061956 |
- |
- |
PREDICTED: uncharacterized protein LOC105649105 [Jatropha curcas] |
| 5 |
Hb_006681_020 |
0.0880121943 |
- |
- |
PREDICTED: peptidyl-prolyl cis-trans isomerase CYP38, chloroplastic [Jatropha curcas] |
| 6 |
Hb_004855_070 |
0.0898703864 |
- |
- |
PLASTID-SPECIFIC RIBOSOMAL protein 4 [Populus trichocarpa] |
| 7 |
Hb_000768_120 |
0.0909381156 |
- |
- |
PREDICTED: RNA polymerase sigma factor sigB [Jatropha curcas] |
| 8 |
Hb_000322_040 |
0.1016412361 |
- |
- |
PREDICTED: lycopene epsilon cyclase, chloroplastic [Jatropha curcas] |
| 9 |
Hb_002400_010 |
0.1046186283 |
- |
- |
PREDICTED: putative L-ascorbate peroxidase 6 [Jatropha curcas] |
| 10 |
Hb_000637_070 |
0.1099128877 |
- |
- |
PREDICTED: uncharacterized protein LOC105650668 [Jatropha curcas] |
| 11 |
Hb_005488_190 |
0.1104712307 |
- |
- |
RNA polymerase sigma factor rpoD, putative [Ricinus communis] |
| 12 |
Hb_000062_480 |
0.114641497 |
- |
- |
PREDICTED: 4-coumarate--CoA ligase-like 9 isoform X1 [Jatropha curcas] |
| 13 |
Hb_003470_100 |
0.1151167932 |
- |
- |
protein binding protein, putative [Ricinus communis] |
| 14 |
Hb_006583_030 |
0.1169788569 |
- |
- |
PREDICTED: SPX and EXS domain-containing protein 1-like isoform X4 [Jatropha curcas] |
| 15 |
Hb_005000_060 |
0.1182931429 |
- |
- |
PREDICTED: uncharacterized protein LOC105637867 [Jatropha curcas] |
| 16 |
Hb_006252_030 |
0.1186519506 |
- |
- |
PREDICTED: uncharacterized protein LOC105638333 [Jatropha curcas] |
| 17 |
Hb_000056_310 |
0.1200461958 |
- |
- |
PREDICTED: uncharacterized protein LOC105630433 [Jatropha curcas] |
| 18 |
Hb_000703_070 |
0.120339463 |
- |
- |
PREDICTED: uncharacterized protein LOC105635704 isoform X1 [Jatropha curcas] |
| 19 |
Hb_126933_020 |
0.1212732081 |
- |
- |
PREDICTED: uncharacterized protein LOC105638525 [Jatropha curcas] |
| 20 |
Hb_001047_180 |
0.1250715697 |
- |
- |
conserved hypothetical protein [Ricinus communis] |