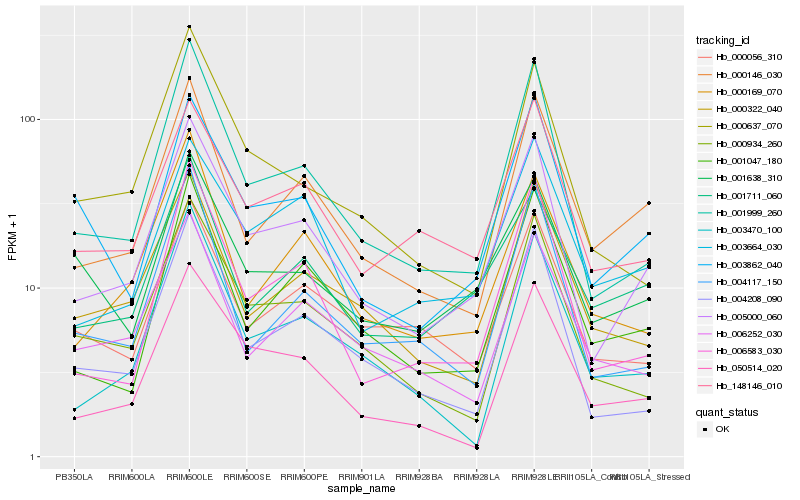
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_005000_060 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105637867 [Jatropha curcas] |
| 2 |
Hb_001999_260 |
0.0934565025 |
- |
- |
PREDICTED: 50S ribosomal protein L4, chloroplastic [Jatropha curcas] |
| 3 |
Hb_000146_030 |
0.114922631 |
- |
- |
PREDICTED: 50S ribosomal protein L10, chloroplastic [Jatropha curcas] |
| 4 |
Hb_148146_010 |
0.1178614841 |
- |
- |
coproporphyrinogen III oxidase [Ziziphus jujuba] |
| 5 |
Hb_004208_090 |
0.1182931429 |
- |
- |
RNA polymerase sigma factor rpoD, putative [Ricinus communis] |
| 6 |
Hb_006583_030 |
0.118726797 |
- |
- |
PREDICTED: SPX and EXS domain-containing protein 1-like isoform X4 [Jatropha curcas] |
| 7 |
Hb_050514_020 |
0.1235468157 |
- |
- |
PREDICTED: abscisic acid receptor PYL4 [Jatropha curcas] |
| 8 |
Hb_003664_030 |
0.1242333167 |
- |
- |
PREDICTED: mitochondrial carnitine/acylcarnitine carrier-like protein [Jatropha curcas] |
| 9 |
Hb_001047_180 |
0.125932308 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 10 |
Hb_000934_260 |
0.1291011871 |
- |
- |
Thylakoid lumenal 29.8 kDa protein, chloroplast precursor, putative [Ricinus communis] |
| 11 |
Hb_003862_040 |
0.1298794098 |
- |
- |
Plastid-specific 30S ribosomal protein 1, chloroplast precursor, putative [Ricinus communis] |
| 12 |
Hb_001711_060 |
0.1338421418 |
- |
- |
PREDICTED: ferredoxin isoform X1 [Jatropha curcas] |
| 13 |
Hb_000322_040 |
0.1350963143 |
- |
- |
PREDICTED: lycopene epsilon cyclase, chloroplastic [Jatropha curcas] |
| 14 |
Hb_000169_070 |
0.1355043156 |
- |
- |
PREDICTED: protein TRANSPORT INHIBITOR RESPONSE 1 [Jatropha curcas] |
| 15 |
Hb_000056_310 |
0.1403308111 |
- |
- |
PREDICTED: uncharacterized protein LOC105630433 [Jatropha curcas] |
| 16 |
Hb_004117_150 |
0.1418592701 |
- |
- |
PREDICTED: uncharacterized protein LOC105649105 [Jatropha curcas] |
| 17 |
Hb_003470_100 |
0.1422951424 |
- |
- |
protein binding protein, putative [Ricinus communis] |
| 18 |
Hb_006252_030 |
0.1437423564 |
- |
- |
PREDICTED: uncharacterized protein LOC105638333 [Jatropha curcas] |
| 19 |
Hb_000637_070 |
0.144471438 |
- |
- |
PREDICTED: uncharacterized protein LOC105650668 [Jatropha curcas] |
| 20 |
Hb_001638_310 |
0.1452679039 |
- |
- |
ribonucleoprotein, chloroplast, putative [Ricinus communis] |