| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
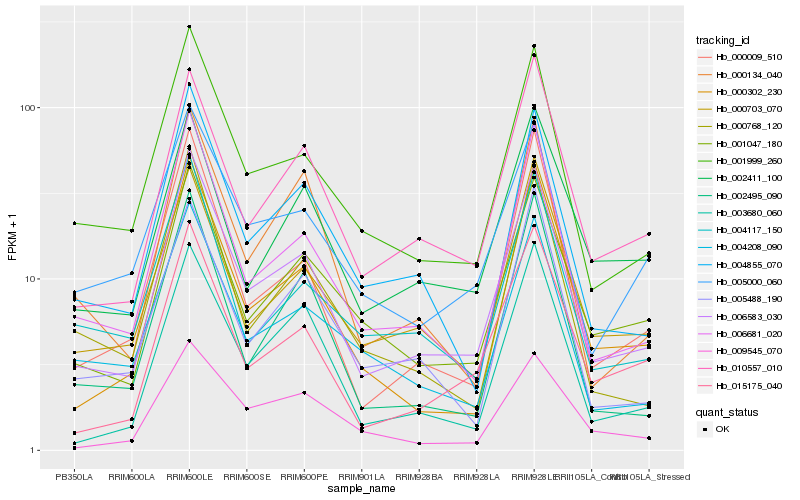
Hb_006583_030 |
0.0 |
- |
- |
PREDICTED: SPX and EXS domain-containing protein 1-like isoform X4 [Jatropha curcas] |
| 2 |
Hb_006681_020 |
0.0809136526 |
- |
- |
PREDICTED: peptidyl-prolyl cis-trans isomerase CYP38, chloroplastic [Jatropha curcas] |
| 3 |
Hb_001999_260 |
0.0813975577 |
- |
- |
PREDICTED: 50S ribosomal protein L4, chloroplastic [Jatropha curcas] |
| 4 |
Hb_004855_070 |
0.0957467068 |
- |
- |
PLASTID-SPECIFIC RIBOSOMAL protein 4 [Populus trichocarpa] |
| 5 |
Hb_000302_230 |
0.1000799986 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_001047_180 |
0.1011517452 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 7 |
Hb_010557_010 |
0.1031013653 |
- |
- |
PREDICTED: 50S ribosomal protein L3, chloroplastic [Jatropha curcas] |
| 8 |
Hb_015175_040 |
0.106409799 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 9 |
Hb_000009_510 |
0.1090532344 |
- |
- |
PREDICTED: uncharacterized protein LOC105126496 [Populus euphratica] |
| 10 |
Hb_003680_060 |
0.1108307517 |
- |
- |
hypothetical protein VITISV_022322 [Vitis vinifera] |
| 11 |
Hb_000703_070 |
0.114766004 |
- |
- |
PREDICTED: uncharacterized protein LOC105635704 isoform X1 [Jatropha curcas] |
| 12 |
Hb_000134_040 |
0.1148558163 |
- |
- |
PREDICTED: 30S ribosomal protein 2, chloroplastic [Jatropha curcas] |
| 13 |
Hb_000768_120 |
0.1152484948 |
- |
- |
PREDICTED: RNA polymerase sigma factor sigB [Jatropha curcas] |
| 14 |
Hb_002495_090 |
0.1165488751 |
transcription factor |
TF Family: bHLH |
PREDICTED: transcription factor bHLH48 isoform X2 [Jatropha curcas] |
| 15 |
Hb_004208_090 |
0.1169788569 |
- |
- |
RNA polymerase sigma factor rpoD, putative [Ricinus communis] |
| 16 |
Hb_005000_060 |
0.118726797 |
- |
- |
PREDICTED: uncharacterized protein LOC105637867 [Jatropha curcas] |
| 17 |
Hb_004117_150 |
0.121666998 |
- |
- |
PREDICTED: uncharacterized protein LOC105649105 [Jatropha curcas] |
| 18 |
Hb_009545_070 |
0.1219806836 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At4g14820 [Jatropha curcas] |
| 19 |
Hb_002411_100 |
0.1241798708 |
- |
- |
PREDICTED: uncharacterized protein LOC105631266 [Jatropha curcas] |
| 20 |
Hb_005488_190 |
0.1261193548 |
- |
- |
RNA polymerase sigma factor rpoD, putative [Ricinus communis] |