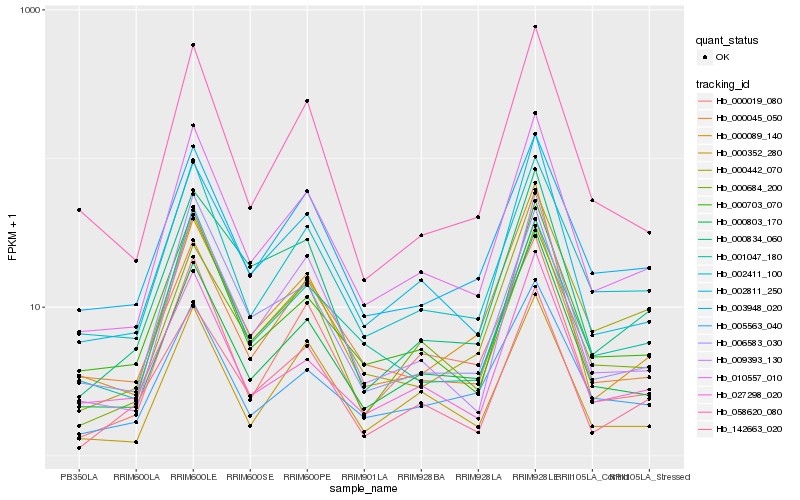
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_010557_010 |
0.0 |
- |
- |
PREDICTED: 50S ribosomal protein L3, chloroplastic [Jatropha curcas] |
| 2 |
Hb_002411_100 |
0.0682247229 |
- |
- |
PREDICTED: uncharacterized protein LOC105631266 [Jatropha curcas] |
| 3 |
Hb_002811_250 |
0.070915222 |
- |
- |
PREDICTED: probable ribose-5-phosphate isomerase 3, chloroplastic [Jatropha curcas] |
| 4 |
Hb_000703_070 |
0.076656051 |
- |
- |
PREDICTED: uncharacterized protein LOC105635704 isoform X1 [Jatropha curcas] |
| 5 |
Hb_000803_170 |
0.078758929 |
- |
- |
PREDICTED: uncharacterized protein LOC105648312 [Jatropha curcas] |
| 6 |
Hb_009393_130 |
0.085319044 |
- |
- |
PREDICTED: uncharacterized protein LOC105638053 isoform X4 [Jatropha curcas] |
| 7 |
Hb_000045_050 |
0.0898392088 |
- |
- |
PREDICTED: probable zinc metalloprotease EGY2, chloroplastic [Jatropha curcas] |
| 8 |
Hb_003948_020 |
0.0904253782 |
- |
- |
PREDICTED: transmembrane protein 41B [Jatropha curcas] |
| 9 |
Hb_000684_200 |
0.0917270618 |
- |
- |
PREDICTED: protein IQ-DOMAIN 1-like [Jatropha curcas] |
| 10 |
Hb_058620_080 |
0.0944714757 |
- |
- |
PREDICTED: phosphoglycerate kinase, chloroplastic [Jatropha curcas] |
| 11 |
Hb_000442_070 |
0.0985640705 |
- |
- |
PREDICTED: uncharacterized protein LOC105645538 [Jatropha curcas] |
| 12 |
Hb_000352_280 |
0.099714074 |
- |
- |
PREDICTED: acetyl-coenzyme A synthetase, chloroplastic/glyoxysomal isoform X2 [Jatropha curcas] |
| 13 |
Hb_001047_180 |
0.1007487534 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 14 |
Hb_027298_020 |
0.1009812126 |
- |
- |
hypothetical protein POPTR_0005s05760g [Populus trichocarpa] |
| 15 |
Hb_000019_080 |
0.1021409446 |
transcription factor |
TF Family: bHLH |
PREDICTED: transcription factor ICE1 [Jatropha curcas] |
| 16 |
Hb_006583_030 |
0.1031013653 |
- |
- |
PREDICTED: SPX and EXS domain-containing protein 1-like isoform X4 [Jatropha curcas] |
| 17 |
Hb_005563_040 |
0.1032933608 |
- |
- |
PREDICTED: uncharacterized protein LOC105630105 isoform X1 [Jatropha curcas] |
| 18 |
Hb_142663_020 |
0.1042700304 |
- |
- |
PREDICTED: uncharacterized protein At1g04910 [Jatropha curcas] |
| 19 |
Hb_000089_140 |
0.1045596019 |
- |
- |
PREDICTED: uncharacterized protein LOC105636682 isoform X2 [Jatropha curcas] |
| 20 |
Hb_000834_060 |
0.1049765911 |
- |
- |
PREDICTED: probable chlorophyll(ide) b reductase NYC1, chloroplastic isoform X1 [Jatropha curcas] |