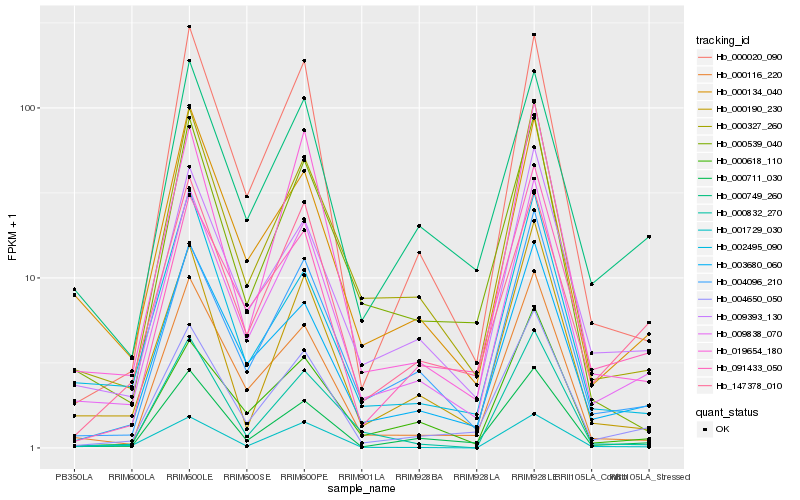
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_009838_070 |
0.0 |
- |
- |
PREDICTED: 28 kDa ribonucleoprotein, chloroplastic [Jatropha curcas] |
| 2 |
Hb_004650_050 |
0.0738683341 |
- |
- |
PREDICTED: uncharacterized protein At1g04910-like isoform X2 [Jatropha curcas] |
| 3 |
Hb_000327_260 |
0.0807223377 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_009393_130 |
0.0816972979 |
- |
- |
PREDICTED: uncharacterized protein LOC105638053 isoform X4 [Jatropha curcas] |
| 5 |
Hb_003680_060 |
0.0848151813 |
- |
- |
hypothetical protein VITISV_022322 [Vitis vinifera] |
| 6 |
Hb_000711_030 |
0.0907802497 |
- |
- |
hypothetical protein POPTR_0019s02860g [Populus trichocarpa] |
| 7 |
Hb_004096_210 |
0.0910562287 |
- |
- |
PREDICTED: uncharacterized protein LOC105638707 isoform X2 [Jatropha curcas] |
| 8 |
Hb_000116_220 |
0.0945129743 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 9 |
Hb_000134_040 |
0.0972792441 |
- |
- |
PREDICTED: 30S ribosomal protein 2, chloroplastic [Jatropha curcas] |
| 10 |
Hb_019654_180 |
0.1064897353 |
- |
- |
glutathione-s-transferase theta, gst, putative [Ricinus communis] |
| 11 |
Hb_000749_260 |
0.1094948816 |
- |
- |
PREDICTED: protochlorophyllide reductase, chloroplastic [Jatropha curcas] |
| 12 |
Hb_000190_230 |
0.1100416499 |
- |
- |
PREDICTED: sulfate transporter 3.1-like [Jatropha curcas] |
| 13 |
Hb_000020_090 |
0.1108931642 |
- |
- |
hypothetical protein JCGZ_09084 [Jatropha curcas] |
| 14 |
Hb_000618_110 |
0.1114794533 |
- |
- |
hypothetical protein POPTR_0003s09870g [Populus trichocarpa] |
| 15 |
Hb_002495_090 |
0.1165798679 |
transcription factor |
TF Family: bHLH |
PREDICTED: transcription factor bHLH48 isoform X2 [Jatropha curcas] |
| 16 |
Hb_147378_010 |
0.1177427711 |
- |
- |
- |
| 17 |
Hb_091433_050 |
0.1182020479 |
- |
- |
PREDICTED: uncharacterized protein LOC105646626 [Jatropha curcas] |
| 18 |
Hb_000539_040 |
0.1182537729 |
- |
- |
PREDICTED: uncharacterized protein LOC105636110 isoform X1 [Jatropha curcas] |
| 19 |
Hb_001729_030 |
0.1185733639 |
- |
- |
hypothetical protein VITISV_044457 [Vitis vinifera] |
| 20 |
Hb_000832_270 |
0.1192741846 |
- |
- |
conserved hypothetical protein [Ricinus communis] |