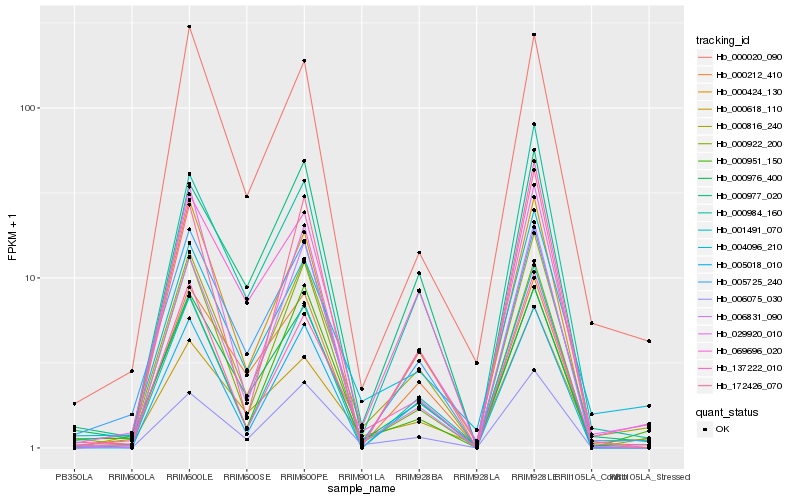
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000816_240 |
0.0 |
- |
- |
PREDICTED: phosphoglucan phosphatase LSF2, chloroplastic [Jatropha curcas] |
| 2 |
Hb_137222_010 |
0.0855855775 |
- |
- |
PREDICTED: hippocampus abundant transcript-like protein 1 [Jatropha curcas] |
| 3 |
Hb_005725_240 |
0.0981153665 |
transcription factor |
TF Family: bHLH |
PREDICTED: transcription factor bHLH74 isoform X1 [Jatropha curcas] |
| 4 |
Hb_004096_210 |
0.1003535075 |
- |
- |
PREDICTED: uncharacterized protein LOC105638707 isoform X2 [Jatropha curcas] |
| 5 |
Hb_000976_400 |
0.1026617503 |
- |
- |
synaptotagmin, putative [Ricinus communis] |
| 6 |
Hb_069696_020 |
0.1063090496 |
- |
- |
PREDICTED: protochlorophyllide-dependent translocon component 52, chloroplastic-like isoform X2 [Jatropha curcas] |
| 7 |
Hb_006075_030 |
0.1098041058 |
- |
- |
PREDICTED: uncharacterized protein LOC105641890 isoform X1 [Jatropha curcas] |
| 8 |
Hb_000424_130 |
0.1105342522 |
- |
- |
PREDICTED: monoacylglycerol lipase abhd6-B-like [Jatropha curcas] |
| 9 |
Hb_005018_010 |
0.1109103792 |
- |
- |
chloroplast-targeted copper chaperone, putative [Ricinus communis] |
| 10 |
Hb_029920_010 |
0.1118175435 |
- |
- |
pepsin A, putative [Ricinus communis] |
| 11 |
Hb_000984_160 |
0.112256577 |
- |
- |
PREDICTED: amino acid permease 6-like isoform X1 [Jatropha curcas] |
| 12 |
Hb_000951_150 |
0.1132141188 |
- |
- |
hypothetical protein POPTR_0015s05850g [Populus trichocarpa] |
| 13 |
Hb_001491_070 |
0.1134458051 |
- |
- |
PREDICTED: probable beta-1,3-galactosyltransferase 17 isoform X1 [Jatropha curcas] |
| 14 |
Hb_000922_200 |
0.1148726872 |
transcription factor |
TF Family: C2C2-Dof |
hypothetical protein POPTR_0012s08280g [Populus trichocarpa] |
| 15 |
Hb_000212_410 |
0.1159732227 |
- |
- |
PREDICTED: serine carboxypeptidase 1-like [Jatropha curcas] |
| 16 |
Hb_000020_090 |
0.1165528399 |
- |
- |
hypothetical protein JCGZ_09084 [Jatropha curcas] |
| 17 |
Hb_000618_110 |
0.1166612697 |
- |
- |
hypothetical protein POPTR_0003s09870g [Populus trichocarpa] |
| 18 |
Hb_000977_020 |
0.1189397605 |
- |
- |
beta-hexosaminidase, putative [Ricinus communis] |
| 19 |
Hb_006831_090 |
0.11994867 |
- |
- |
PREDICTED: uncharacterized protein LOC105642125 isoform X1 [Jatropha curcas] |
| 20 |
Hb_172426_070 |
0.1206850931 |
- |
- |
PREDICTED: BAG family molecular chaperone regulator 3-like isoform X1 [Jatropha curcas] |