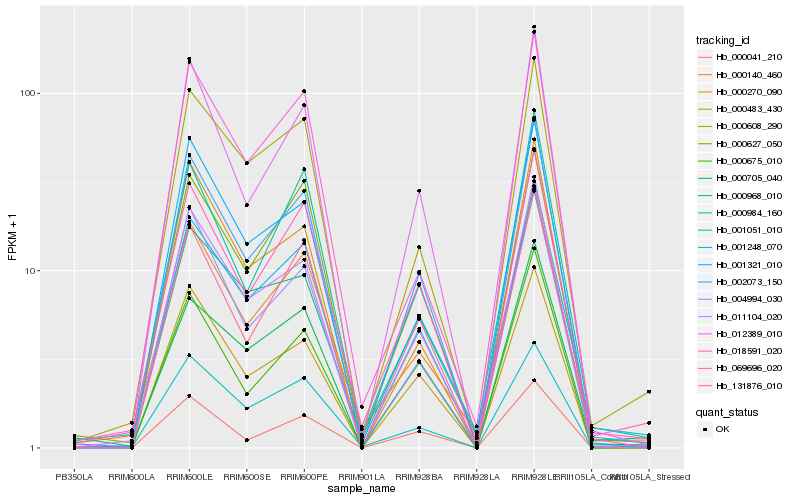
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002073_150 |
0.0 |
transcription factor |
TF Family: MYB-related |
PREDICTED: transcription factor MYB1R1 [Jatropha curcas] |
| 2 |
Hb_011104_020 |
0.0489993042 |
- |
- |
PREDICTED: ABC transporter B family member 2-like [Jatropha curcas] |
| 3 |
Hb_012389_010 |
0.0492835469 |
- |
- |
hypothetical protein POPTR_0018s11820g [Populus trichocarpa] |
| 4 |
Hb_001248_070 |
0.050866893 |
- |
- |
PREDICTED: probable polyamine transporter At3g19553 [Jatropha curcas] |
| 5 |
Hb_131876_010 |
0.0634512462 |
transcription factor |
TF Family: MYB |
PREDICTED: myb-related protein 308-like isoform X1 [Jatropha curcas] |
| 6 |
Hb_000483_430 |
0.0662189221 |
- |
- |
- |
| 7 |
Hb_069696_020 |
0.0706791983 |
- |
- |
PREDICTED: protochlorophyllide-dependent translocon component 52, chloroplastic-like isoform X2 [Jatropha curcas] |
| 8 |
Hb_004994_030 |
0.0759387879 |
transcription factor |
TF Family: HB |
PREDICTED: homeobox-leucine zipper protein ANTHOCYANINLESS 2 [Jatropha curcas] |
| 9 |
Hb_000140_460 |
0.0775846501 |
- |
- |
PREDICTED: uncharacterized protein LOC105628417 [Jatropha curcas] |
| 10 |
Hb_000041_210 |
0.0802018868 |
- |
- |
PREDICTED: cysteine-rich receptor-like protein kinase 42 [Jatropha curcas] |
| 11 |
Hb_000608_290 |
0.0826056015 |
- |
- |
thioredoxin f-type, putative [Ricinus communis] |
| 12 |
Hb_001051_010 |
0.0831033591 |
- |
- |
PREDICTED: putative cyclic nucleotide-gated ion channel 15 [Jatropha curcas] |
| 13 |
Hb_000705_040 |
0.0836759705 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 14 |
Hb_000984_160 |
0.0844202892 |
- |
- |
PREDICTED: amino acid permease 6-like isoform X1 [Jatropha curcas] |
| 15 |
Hb_000675_010 |
0.0846105182 |
- |
- |
PREDICTED: cytokinin dehydrogenase 7 [Jatropha curcas] |
| 16 |
Hb_001321_010 |
0.0864275822 |
- |
- |
HIGH-CHLOROPHYLL-FLUORESCENCE 101 family protein [Populus trichocarpa] |
| 17 |
Hb_000270_090 |
0.0891853563 |
- |
- |
nitrate transporter, putative [Ricinus communis] |
| 18 |
Hb_000627_050 |
0.0896292678 |
- |
- |
PREDICTED: uncharacterized protein LOC105632845 [Jatropha curcas] |
| 19 |
Hb_000968_010 |
0.0959014404 |
- |
- |
PREDICTED: glutamate receptor 3.7 isoform X1 [Jatropha curcas] |
| 20 |
Hb_018591_020 |
0.0965721634 |
- |
- |
Ribulose bisphosphate carboxylase/oxygenase activase 1, chloroplast precursor, putative [Ricinus communis] |