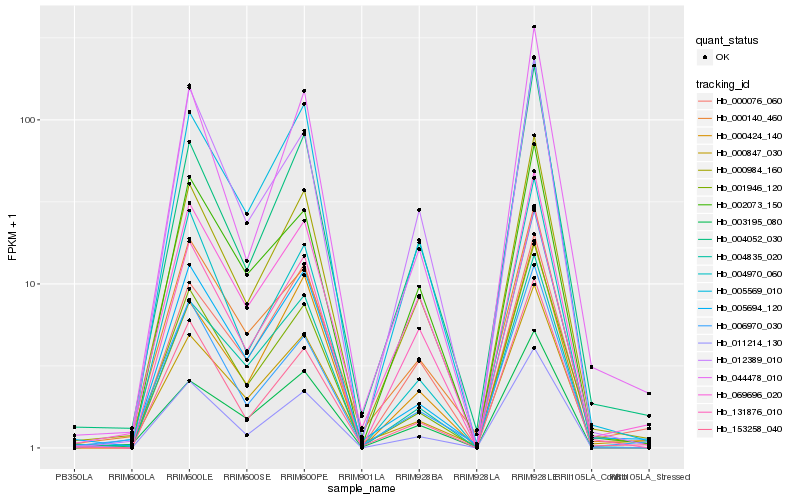
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000984_160 |
0.0 |
- |
- |
PREDICTED: amino acid permease 6-like isoform X1 [Jatropha curcas] |
| 2 |
Hb_005569_010 |
0.0533349119 |
- |
- |
Heme-binding protein, putative [Ricinus communis] |
| 3 |
Hb_131876_010 |
0.0692518747 |
transcription factor |
TF Family: MYB |
PREDICTED: myb-related protein 308-like isoform X1 [Jatropha curcas] |
| 4 |
Hb_004052_030 |
0.0704709089 |
- |
- |
PREDICTED: uncharacterized protein LOC105640637 [Jatropha curcas] |
| 5 |
Hb_069696_020 |
0.0735891057 |
- |
- |
PREDICTED: protochlorophyllide-dependent translocon component 52, chloroplastic-like isoform X2 [Jatropha curcas] |
| 6 |
Hb_001946_120 |
0.0767863727 |
- |
- |
PREDICTED: SPX domain-containing protein 1-like [Jatropha curcas] |
| 7 |
Hb_000847_030 |
0.079111447 |
- |
- |
PREDICTED: asparagine--tRNA ligase, cytoplasmic 2 [Jatropha curcas] |
| 8 |
Hb_011214_130 |
0.0804685488 |
transcription factor |
TF Family: NAC |
PREDICTED: NAC domain-containing protein 86 [Jatropha curcas] |
| 9 |
Hb_005694_120 |
0.0811426286 |
- |
- |
PREDICTED: calcium-transporting ATPase, endoplasmic reticulum-type [Jatropha curcas] |
| 10 |
Hb_000140_460 |
0.0817714853 |
- |
- |
PREDICTED: uncharacterized protein LOC105628417 [Jatropha curcas] |
| 11 |
Hb_004835_020 |
0.0828363784 |
- |
- |
PREDICTED: receptor-like protein 12 [Populus euphratica] |
| 12 |
Hb_044478_010 |
0.0833375888 |
- |
- |
plastoquinol-plastocyanin reductase, putative [Ricinus communis] |
| 13 |
Hb_002073_150 |
0.0844202892 |
transcription factor |
TF Family: MYB-related |
PREDICTED: transcription factor MYB1R1 [Jatropha curcas] |
| 14 |
Hb_003195_080 |
0.0848899035 |
- |
- |
PREDICTED: cytochrome P450 87A3-like [Jatropha curcas] |
| 15 |
Hb_012389_010 |
0.0849833811 |
- |
- |
hypothetical protein POPTR_0018s11820g [Populus trichocarpa] |
| 16 |
Hb_153258_040 |
0.092225659 |
- |
- |
PREDICTED: type I inositol 1,4,5-trisphosphate 5-phosphatase CVP2-like [Jatropha curcas] |
| 17 |
Hb_000424_140 |
0.0926072145 |
- |
- |
Protein kinase APK1A, chloroplast precursor, putative [Ricinus communis] |
| 18 |
Hb_000076_060 |
0.0942022158 |
- |
- |
hypothetical protein RCOM_0586940 [Ricinus communis] |
| 19 |
Hb_006970_030 |
0.0960644864 |
- |
- |
synaptotagmin, putative [Ricinus communis] |
| 20 |
Hb_004970_060 |
0.0970384773 |
- |
- |
PREDICTED: uncharacterized protein LOC100259061 [Vitis vinifera] |