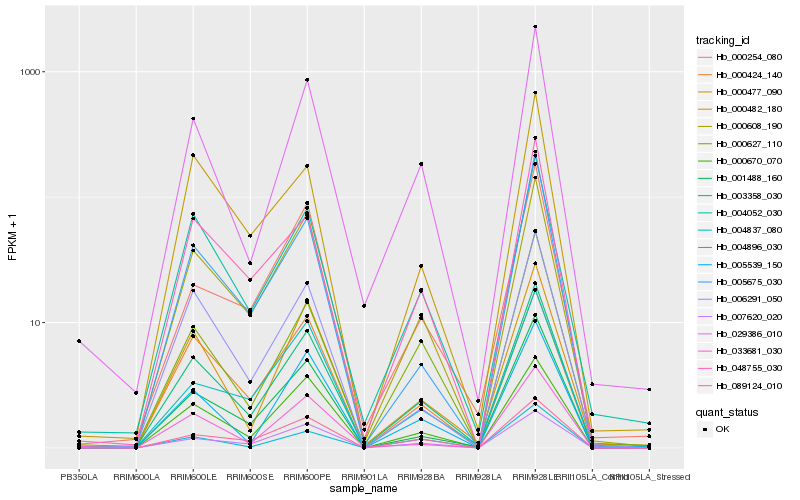
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_003358_030 |
0.0 |
- |
- |
ankyrin-kinase, putative [Ricinus communis] |
| 2 |
Hb_000608_190 |
0.0712221626 |
rubber biosynthesis |
Gene Name: Isopentenyl-diphosphate delta isomerase |
PREDICTED: isopentenyl-diphosphate Delta-isomerase I [Jatropha curcas] |
| 3 |
Hb_029386_010 |
0.0727565338 |
- |
- |
RALF-LIKE 27 family protein [Populus trichocarpa] |
| 4 |
Hb_004052_030 |
0.0829168498 |
- |
- |
PREDICTED: uncharacterized protein LOC105640637 [Jatropha curcas] |
| 5 |
Hb_001488_160 |
0.0876235003 |
- |
- |
UDP-glucuronosyltransferase, putative [Ricinus communis] |
| 6 |
Hb_000477_090 |
0.0914790834 |
- |
- |
PREDICTED: uncharacterized protein LOC105639302 [Jatropha curcas] |
| 7 |
Hb_007620_020 |
0.0942383426 |
- |
- |
PREDICTED: cationic amino acid transporter 7, chloroplastic-like isoform X1 [Jatropha curcas] |
| 8 |
Hb_000670_070 |
0.0981766663 |
- |
- |
PREDICTED: probable carbohydrate esterase At4g34215 [Jatropha curcas] |
| 9 |
Hb_048755_030 |
0.0981961923 |
- |
- |
Periplasmic beta-glucosidase precursor, putative [Ricinus communis] |
| 10 |
Hb_089124_010 |
0.1020276688 |
- |
- |
hypothetical protein POPTR_0018s09540g [Populus trichocarpa] |
| 11 |
Hb_033681_030 |
0.1065670641 |
desease resistance |
Gene Name: NB-ARC |
Leucine-rich repeat containing protein [Theobroma cacao] |
| 12 |
Hb_005675_030 |
0.1065818398 |
- |
- |
PREDICTED: 3-ketoacyl-CoA synthase 6 [Jatropha curcas] |
| 13 |
Hb_005539_150 |
0.1073580885 |
- |
- |
PREDICTED: probable xyloglucan endotransglucosylase/hydrolase protein 33 [Jatropha curcas] |
| 14 |
Hb_000627_110 |
0.108213472 |
- |
- |
cellulose synthase, putative [Ricinus communis] |
| 15 |
Hb_006291_050 |
0.1086987875 |
- |
- |
PREDICTED: polyribonucleotide nucleotidyltransferase 2, mitochondrial isoform X1 [Jatropha curcas] |
| 16 |
Hb_004896_030 |
0.1141704294 |
- |
- |
hypothetical protein VITISV_007475 [Vitis vinifera] |
| 17 |
Hb_004837_080 |
0.1150961903 |
transcription factor |
TF Family: MYB |
PREDICTED: transcription factor MYB46 [Jatropha curcas] |
| 18 |
Hb_000424_140 |
0.1156418715 |
- |
- |
Protein kinase APK1A, chloroplast precursor, putative [Ricinus communis] |
| 19 |
Hb_000482_180 |
0.1166348176 |
- |
- |
PREDICTED: glycine dehydrogenase (decarboxylating), mitochondrial [Jatropha curcas] |
| 20 |
Hb_000254_080 |
0.1171732133 |
- |
- |
PREDICTED: uncharacterized protein LOC105635000 [Jatropha curcas] |