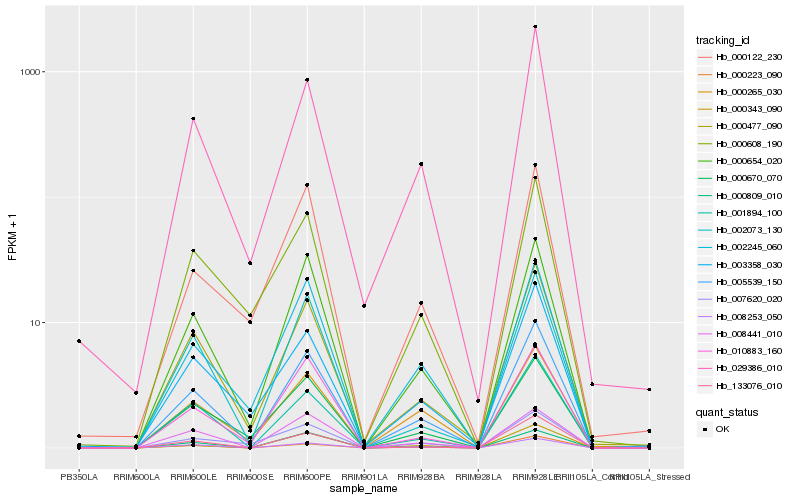
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_005539_150 |
0.0 |
- |
- |
PREDICTED: probable xyloglucan endotransglucosylase/hydrolase protein 33 [Jatropha curcas] |
| 2 |
Hb_000477_090 |
0.0730682431 |
- |
- |
PREDICTED: uncharacterized protein LOC105639302 [Jatropha curcas] |
| 3 |
Hb_000654_020 |
0.0734788235 |
- |
- |
hypothetical protein CICLE_v10006262mg [Citrus clementina] |
| 4 |
Hb_029386_010 |
0.0887839222 |
- |
- |
RALF-LIKE 27 family protein [Populus trichocarpa] |
| 5 |
Hb_002245_060 |
0.089679339 |
- |
- |
PREDICTED: kinesin-3 isoform X1 [Jatropha curcas] |
| 6 |
Hb_008253_050 |
0.0905192481 |
transcription factor |
TF Family: MYB |
hypothetical protein POPTR_0001s36220g [Populus trichocarpa] |
| 7 |
Hb_000670_070 |
0.0965370581 |
- |
- |
PREDICTED: probable carbohydrate esterase At4g34215 [Jatropha curcas] |
| 8 |
Hb_010883_160 |
0.1003366875 |
- |
- |
PREDICTED: pathogenesis-related protein 5 isoform X1 [Jatropha curcas] |
| 9 |
Hb_000265_030 |
0.103668733 |
- |
- |
PREDICTED: subtilisin-like protease SBT5.3 [Jatropha curcas] |
| 10 |
Hb_001894_100 |
0.1064697656 |
- |
- |
PREDICTED: kininogen-1-like [Jatropha curcas] |
| 11 |
Hb_003358_030 |
0.1073580885 |
- |
- |
ankyrin-kinase, putative [Ricinus communis] |
| 12 |
Hb_002073_130 |
0.1086949177 |
- |
- |
PREDICTED: uncharacterized protein LOC100246830 [Vitis vinifera] |
| 13 |
Hb_000223_090 |
0.1102898551 |
- |
- |
Blue copper protein precursor, putative [Ricinus communis] |
| 14 |
Hb_133076_010 |
0.1126136624 |
- |
- |
PREDICTED: scopoletin glucosyltransferase-like [Populus euphratica] |
| 15 |
Hb_008441_010 |
0.1140874812 |
- |
- |
leucine-rich repeat receptor protein kinase exs precursor, putative [Ricinus communis] |
| 16 |
Hb_000809_010 |
0.1151832279 |
- |
- |
hypothetical protein JCGZ_12342 [Jatropha curcas] |
| 17 |
Hb_007620_020 |
0.1168187764 |
- |
- |
PREDICTED: cationic amino acid transporter 7, chloroplastic-like isoform X1 [Jatropha curcas] |
| 18 |
Hb_000122_230 |
0.1172622242 |
- |
- |
laccase, putative [Ricinus communis] |
| 19 |
Hb_000608_190 |
0.1180404356 |
rubber biosynthesis |
Gene Name: Isopentenyl-diphosphate delta isomerase |
PREDICTED: isopentenyl-diphosphate Delta-isomerase I [Jatropha curcas] |
| 20 |
Hb_000343_090 |
0.1184931888 |
transcription factor |
TF Family: OFP |
conserved hypothetical protein [Ricinus communis] |