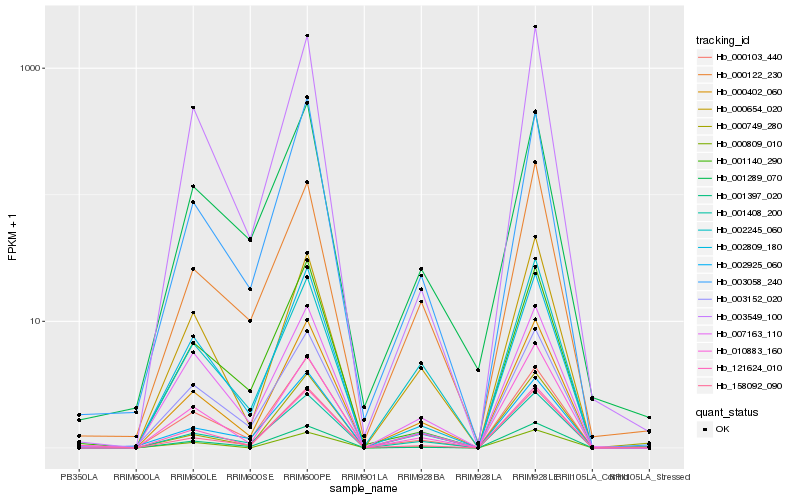
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000402_060 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105644642 isoform X2 [Jatropha curcas] |
| 2 |
Hb_001408_200 |
0.0513105982 |
- |
- |
hypothetical protein JCGZ_08964 [Jatropha curcas] |
| 3 |
Hb_010883_160 |
0.0712323007 |
- |
- |
PREDICTED: pathogenesis-related protein 5 isoform X1 [Jatropha curcas] |
| 4 |
Hb_003058_240 |
0.074749976 |
- |
- |
PREDICTED: tubulin beta-5 chain-like [Jatropha curcas] |
| 5 |
Hb_000654_020 |
0.0831265519 |
- |
- |
hypothetical protein CICLE_v10006262mg [Citrus clementina] |
| 6 |
Hb_002809_180 |
0.0857215618 |
- |
- |
PREDICTED: homeobox protein HD1 [Jatropha curcas] |
| 7 |
Hb_001397_020 |
0.0865095749 |
- |
- |
cytochrome P450 family protein [Populus trichocarpa] |
| 8 |
Hb_003152_020 |
0.0884325965 |
- |
- |
PREDICTED: neutral alpha-glucosidase C [Jatropha curcas] |
| 9 |
Hb_158092_090 |
0.0941859038 |
- |
- |
zinc finger protein, putative [Ricinus communis] |
| 10 |
Hb_000103_440 |
0.0946183204 |
- |
- |
hypothetical protein POPTR_0008s15900g [Populus trichocarpa] |
| 11 |
Hb_002925_060 |
0.0950937655 |
desease resistance |
Gene Name: NB-ARC |
hypothetical protein JCGZ_01547 [Jatropha curcas] |
| 12 |
Hb_000122_230 |
0.0975698718 |
- |
- |
laccase, putative [Ricinus communis] |
| 13 |
Hb_002245_060 |
0.0995604893 |
- |
- |
PREDICTED: kinesin-3 isoform X1 [Jatropha curcas] |
| 14 |
Hb_000809_010 |
0.1016672335 |
- |
- |
hypothetical protein JCGZ_12342 [Jatropha curcas] |
| 15 |
Hb_003549_100 |
0.1018137383 |
- |
- |
PREDICTED: uncharacterized protein LOC105647410 [Jatropha curcas] |
| 16 |
Hb_001289_070 |
0.103806037 |
- |
- |
PREDICTED: protein YLS9-like [Jatropha curcas] |
| 17 |
Hb_007163_110 |
0.105819476 |
- |
- |
PREDICTED: kinesin-like protein KIF19 [Jatropha curcas] |
| 18 |
Hb_000749_280 |
0.1072909347 |
- |
- |
PREDICTED: uncharacterized protein LOC105138169 [Populus euphratica] |
| 19 |
Hb_121624_010 |
0.1072961979 |
- |
- |
PREDICTED: uncharacterized protein LOC105634410 [Jatropha curcas] |
| 20 |
Hb_001140_290 |
0.1081509962 |
- |
- |
hypothetical protein JCGZ_26107 [Jatropha curcas] |