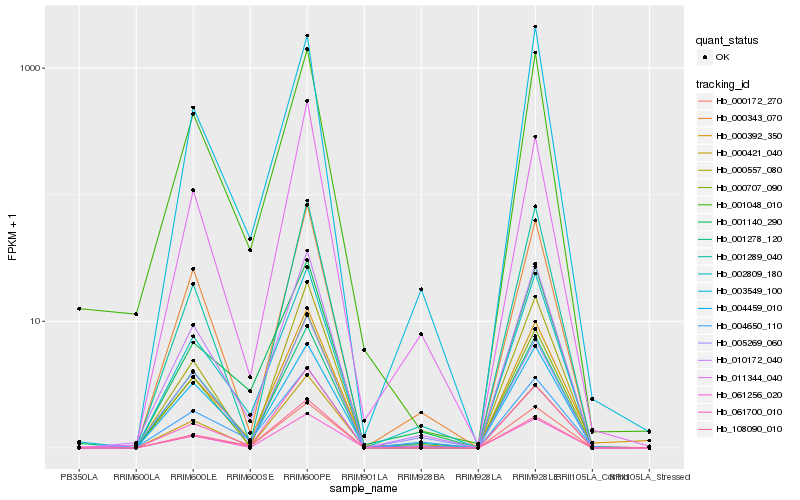
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_010172_040 |
0.0 |
- |
- |
PREDICTED: BAG family molecular chaperone regulator 3-like isoform X1 [Jatropha curcas] |
| 2 |
Hb_002809_180 |
0.0521312569 |
- |
- |
PREDICTED: homeobox protein HD1 [Jatropha curcas] |
| 3 |
Hb_000343_070 |
0.0633682465 |
- |
- |
hypothetical protein JCGZ_06265 [Jatropha curcas] |
| 4 |
Hb_000421_040 |
0.0642225186 |
- |
- |
Peptide transporter, putative [Ricinus communis] |
| 5 |
Hb_000172_270 |
0.0665156075 |
- |
- |
hypothetical protein JCGZ_24087 [Jatropha curcas] |
| 6 |
Hb_000707_090 |
0.0696080115 |
- |
- |
PREDICTED: protein NRT1/ PTR FAMILY 5.6-like [Jatropha curcas] |
| 7 |
Hb_001278_120 |
0.0706239066 |
- |
- |
PREDICTED: reticulon-like protein B14 [Jatropha curcas] |
| 8 |
Hb_061700_010 |
0.0711397018 |
- |
- |
PREDICTED: putative UDP-rhamnose:rhamnosyltransferase 1 [Prunus mume] |
| 9 |
Hb_005269_060 |
0.0776007285 |
- |
- |
PREDICTED: rac-like GTP-binding protein RAC2 [Cicer arietinum] |
| 10 |
Hb_061256_020 |
0.0791073621 |
- |
- |
PREDICTED: UPF0392 protein RCOM_0530710 [Jatropha curcas] |
| 11 |
Hb_001048_010 |
0.0802512738 |
- |
- |
PREDICTED: probable xyloglucan endotransglucosylase/hydrolase protein 7 [Jatropha curcas] |
| 12 |
Hb_000392_350 |
0.0816328418 |
- |
- |
zinc finger protein, putative [Ricinus communis] |
| 13 |
Hb_004459_010 |
0.0835933077 |
- |
- |
PREDICTED: phospholipase A1-IIgamma-like [Jatropha curcas] |
| 14 |
Hb_001289_040 |
0.0850647513 |
- |
- |
NDR1/HIN1-like 1 [Theobroma cacao] |
| 15 |
Hb_000557_080 |
0.0858516978 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 16 |
Hb_001140_290 |
0.0872541801 |
- |
- |
hypothetical protein JCGZ_26107 [Jatropha curcas] |
| 17 |
Hb_003549_100 |
0.088100604 |
- |
- |
PREDICTED: uncharacterized protein LOC105647410 [Jatropha curcas] |
| 18 |
Hb_004650_110 |
0.0881212786 |
- |
- |
hypothetical protein POPTR_0002s08760g [Populus trichocarpa] |
| 19 |
Hb_108090_010 |
0.0896902094 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 20 |
Hb_011344_040 |
0.0942538782 |
- |
- |
PREDICTED: 21 kDa protein [Jatropha curcas] |