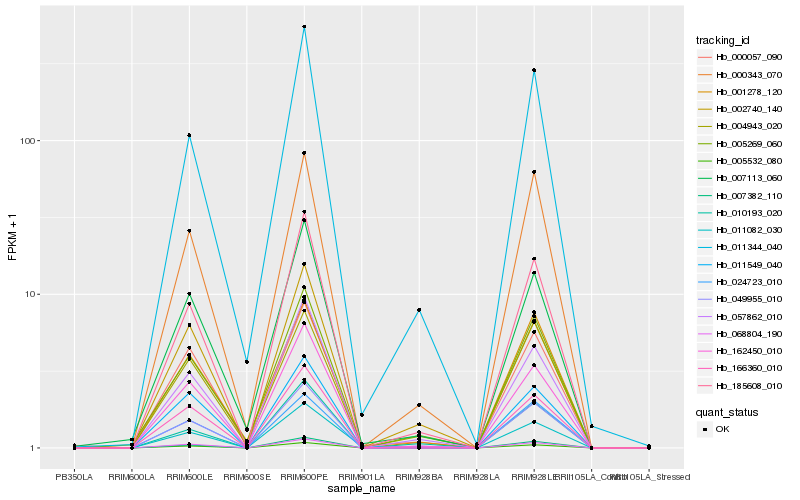
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_049955_010 |
0.0 |
- |
- |
hypothetical protein EUGRSUZ_B00742 [Eucalyptus grandis] |
| 2 |
Hb_185608_010 |
0.0545400462 |
- |
- |
hypothetical protein POPTR_0004s22210g [Populus trichocarpa] |
| 3 |
Hb_162450_010 |
0.0608235445 |
- |
- |
PREDICTED: glucan endo-1,3-beta-glucosidase 12 [Jatropha curcas] |
| 4 |
Hb_005269_060 |
0.0614958653 |
- |
- |
PREDICTED: rac-like GTP-binding protein RAC2 [Cicer arietinum] |
| 5 |
Hb_057862_010 |
0.0623475118 |
- |
- |
protein binding protein, putative [Ricinus communis] |
| 6 |
Hb_000343_070 |
0.0629125805 |
- |
- |
hypothetical protein JCGZ_06265 [Jatropha curcas] |
| 7 |
Hb_005532_080 |
0.0718543217 |
- |
- |
PREDICTED: heparanase-like protein 2 [Jatropha curcas] |
| 8 |
Hb_007382_110 |
0.072408743 |
- |
- |
PREDICTED: bidirectional sugar transporter N3 [Jatropha curcas] |
| 9 |
Hb_011344_040 |
0.078582985 |
- |
- |
PREDICTED: 21 kDa protein [Jatropha curcas] |
| 10 |
Hb_004943_020 |
0.0794841658 |
- |
- |
cationic amino acid transporter, putative [Ricinus communis] |
| 11 |
Hb_166360_010 |
0.0800808773 |
- |
- |
hypothetical protein POPTR_0010s12130g [Populus trichocarpa] |
| 12 |
Hb_068804_190 |
0.0807372236 |
- |
- |
kinase, putative [Ricinus communis] |
| 13 |
Hb_010193_020 |
0.0841774458 |
- |
- |
PREDICTED: NADH--cytochrome b5 reductase 1-like [Jatropha curcas] |
| 14 |
Hb_000057_090 |
0.0842673049 |
- |
- |
hypothetical protein RCOM_1494490 [Ricinus communis] |
| 15 |
Hb_001278_120 |
0.0867850698 |
- |
- |
PREDICTED: reticulon-like protein B14 [Jatropha curcas] |
| 16 |
Hb_011082_030 |
0.0879175972 |
- |
- |
hypothetical protein RCOM_0852210 [Ricinus communis] |
| 17 |
Hb_002740_140 |
0.090534712 |
- |
- |
hypothetical protein L484_022679 [Morus notabilis] |
| 18 |
Hb_007113_060 |
0.0948210168 |
- |
- |
PREDICTED: rac-like GTP-binding protein RAC2 [Jatropha curcas] |
| 19 |
Hb_011549_040 |
0.0965523767 |
- |
- |
- |
| 20 |
Hb_024723_010 |
0.0970406275 |
- |
- |
PREDICTED: leucine-rich repeat receptor protein kinase EMS1-like [Jatropha curcas] |