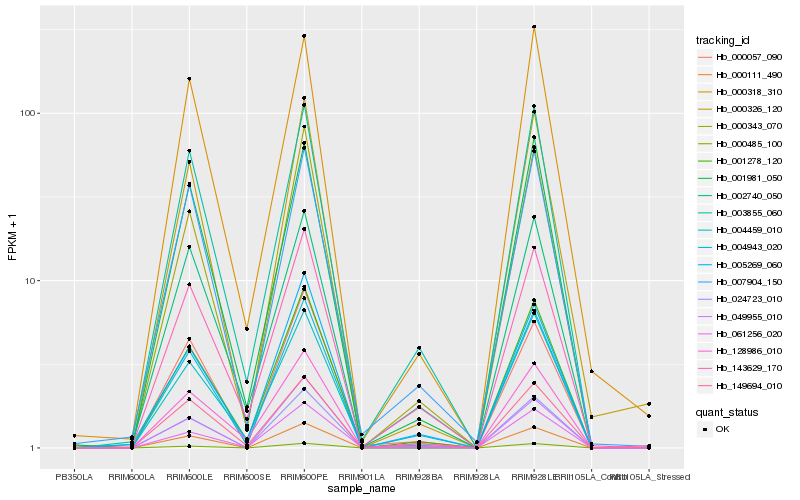
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_004943_020 |
0.0 |
- |
- |
cationic amino acid transporter, putative [Ricinus communis] |
| 2 |
Hb_001278_120 |
0.0437098292 |
- |
- |
PREDICTED: reticulon-like protein B14 [Jatropha curcas] |
| 3 |
Hb_003855_060 |
0.0516264736 |
- |
- |
PREDICTED: uncharacterized protein At4g06744-like [Jatropha curcas] |
| 4 |
Hb_000343_070 |
0.0550498682 |
- |
- |
hypothetical protein JCGZ_06265 [Jatropha curcas] |
| 5 |
Hb_143629_170 |
0.0590664565 |
- |
- |
PREDICTED: pectin acetylesterase 12 [Jatropha curcas] |
| 6 |
Hb_007904_150 |
0.0739263922 |
- |
- |
PREDICTED: probable pectate lyase 5 [Populus euphratica] |
| 7 |
Hb_004459_010 |
0.074019061 |
- |
- |
PREDICTED: phospholipase A1-IIgamma-like [Jatropha curcas] |
| 8 |
Hb_005269_060 |
0.0749861326 |
- |
- |
PREDICTED: rac-like GTP-binding protein RAC2 [Cicer arietinum] |
| 9 |
Hb_002740_050 |
0.0761942279 |
transcription factor |
TF Family: MYB |
PREDICTED: transcription factor MYB86-like [Populus euphratica] |
| 10 |
Hb_128986_010 |
0.0764789924 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_061256_020 |
0.0766139997 |
- |
- |
PREDICTED: UPF0392 protein RCOM_0530710 [Jatropha curcas] |
| 12 |
Hb_001981_050 |
0.0791802064 |
- |
- |
PREDICTED: aldehyde dehydrogenase family 3 member F1-like [Jatropha curcas] |
| 13 |
Hb_000326_120 |
0.0793692901 |
transcription factor |
TF Family: MYB-related |
transcription factor, putative [Ricinus communis] |
| 14 |
Hb_049955_010 |
0.0794841658 |
- |
- |
hypothetical protein EUGRSUZ_B00742 [Eucalyptus grandis] |
| 15 |
Hb_024723_010 |
0.0865598454 |
- |
- |
PREDICTED: leucine-rich repeat receptor protein kinase EMS1-like [Jatropha curcas] |
| 16 |
Hb_149694_010 |
0.086887964 |
- |
- |
Xylem serine proteinase 1 precursor, putative [Ricinus communis] |
| 17 |
Hb_000111_490 |
0.087901629 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_000057_090 |
0.0900193409 |
- |
- |
hypothetical protein RCOM_1494490 [Ricinus communis] |
| 19 |
Hb_000318_310 |
0.0915072954 |
- |
- |
hypothetical protein POPTR_0009s11190g [Populus trichocarpa] |
| 20 |
Hb_000485_100 |
0.0930823002 |
- |
- |
PREDICTED: putative UPF0481 protein At3g02645 [Populus euphratica] |