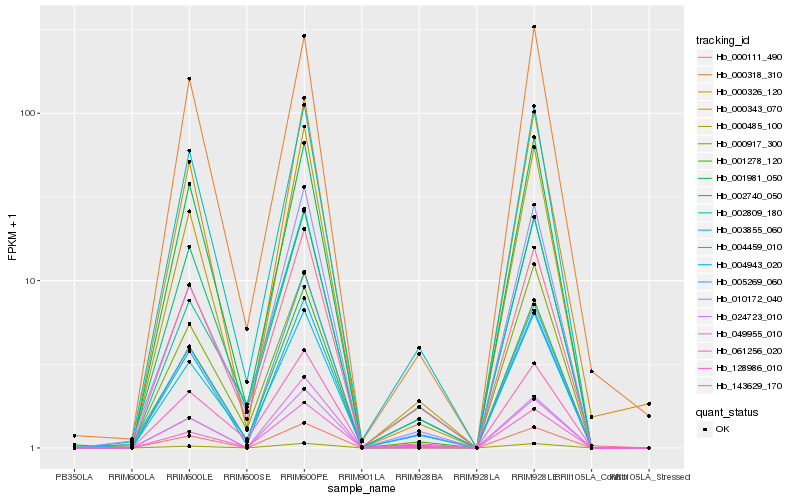
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001278_120 |
0.0 |
- |
- |
PREDICTED: reticulon-like protein B14 [Jatropha curcas] |
| 2 |
Hb_000343_070 |
0.0423520551 |
- |
- |
hypothetical protein JCGZ_06265 [Jatropha curcas] |
| 3 |
Hb_004943_020 |
0.0437098292 |
- |
- |
cationic amino acid transporter, putative [Ricinus communis] |
| 4 |
Hb_004459_010 |
0.0460374731 |
- |
- |
PREDICTED: phospholipase A1-IIgamma-like [Jatropha curcas] |
| 5 |
Hb_128986_010 |
0.0631965051 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_003855_060 |
0.0659822639 |
- |
- |
PREDICTED: uncharacterized protein At4g06744-like [Jatropha curcas] |
| 7 |
Hb_000326_120 |
0.0697782333 |
transcription factor |
TF Family: MYB-related |
transcription factor, putative [Ricinus communis] |
| 8 |
Hb_143629_170 |
0.0700800696 |
- |
- |
PREDICTED: pectin acetylesterase 12 [Jatropha curcas] |
| 9 |
Hb_010172_040 |
0.0706239066 |
- |
- |
PREDICTED: BAG family molecular chaperone regulator 3-like isoform X1 [Jatropha curcas] |
| 10 |
Hb_005269_060 |
0.0720669167 |
- |
- |
PREDICTED: rac-like GTP-binding protein RAC2 [Cicer arietinum] |
| 11 |
Hb_001981_050 |
0.0746359071 |
- |
- |
PREDICTED: aldehyde dehydrogenase family 3 member F1-like [Jatropha curcas] |
| 12 |
Hb_061256_020 |
0.0751862587 |
- |
- |
PREDICTED: UPF0392 protein RCOM_0530710 [Jatropha curcas] |
| 13 |
Hb_002809_180 |
0.0756501507 |
- |
- |
PREDICTED: homeobox protein HD1 [Jatropha curcas] |
| 14 |
Hb_024723_010 |
0.0759554293 |
- |
- |
PREDICTED: leucine-rich repeat receptor protein kinase EMS1-like [Jatropha curcas] |
| 15 |
Hb_000111_490 |
0.0797057405 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 16 |
Hb_000485_100 |
0.0801598069 |
- |
- |
PREDICTED: putative UPF0481 protein At3g02645 [Populus euphratica] |
| 17 |
Hb_000917_300 |
0.0809014641 |
transcription factor |
TF Family: bHLH |
PREDICTED: transcription factor bHLH30-like [Populus euphratica] |
| 18 |
Hb_002740_050 |
0.0863315394 |
transcription factor |
TF Family: MYB |
PREDICTED: transcription factor MYB86-like [Populus euphratica] |
| 19 |
Hb_049955_010 |
0.0867850698 |
- |
- |
hypothetical protein EUGRSUZ_B00742 [Eucalyptus grandis] |
| 20 |
Hb_000318_310 |
0.0870214239 |
- |
- |
hypothetical protein POPTR_0009s11190g [Populus trichocarpa] |