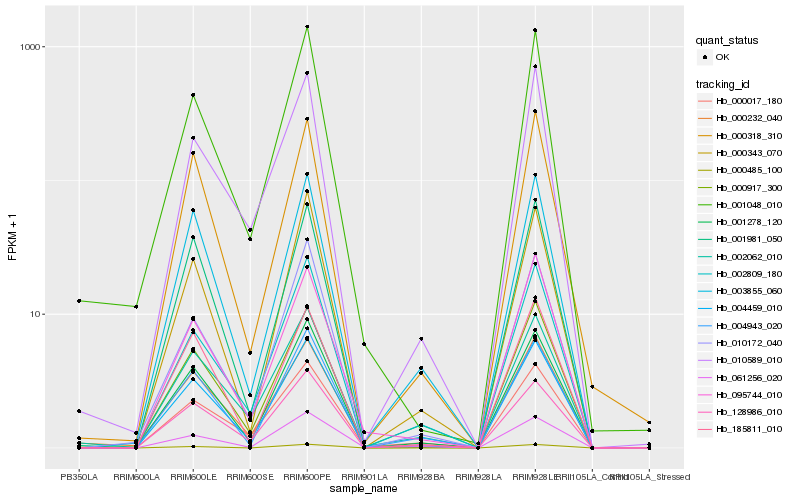
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_004459_010 |
0.0 |
- |
- |
PREDICTED: phospholipase A1-IIgamma-like [Jatropha curcas] |
| 2 |
Hb_001278_120 |
0.0460374731 |
- |
- |
PREDICTED: reticulon-like protein B14 [Jatropha curcas] |
| 3 |
Hb_000917_300 |
0.0537112248 |
transcription factor |
TF Family: bHLH |
PREDICTED: transcription factor bHLH30-like [Populus euphratica] |
| 4 |
Hb_001981_050 |
0.0627370265 |
- |
- |
PREDICTED: aldehyde dehydrogenase family 3 member F1-like [Jatropha curcas] |
| 5 |
Hb_003855_060 |
0.0664687202 |
- |
- |
PREDICTED: uncharacterized protein At4g06744-like [Jatropha curcas] |
| 6 |
Hb_095744_010 |
0.0719262588 |
- |
- |
PREDICTED: G-type lectin S-receptor-like serine/threonine-protein kinase SD1-1 [Fragaria vesca subsp. vesca] |
| 7 |
Hb_000318_310 |
0.0721486416 |
- |
- |
hypothetical protein POPTR_0009s11190g [Populus trichocarpa] |
| 8 |
Hb_004943_020 |
0.074019061 |
- |
- |
cationic amino acid transporter, putative [Ricinus communis] |
| 9 |
Hb_128986_010 |
0.0750916118 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 10 |
Hb_000017_180 |
0.0755730725 |
- |
- |
hypothetical protein JCGZ_20551 [Jatropha curcas] |
| 11 |
Hb_185811_010 |
0.0763166138 |
- |
- |
Flavonol 4'-sulfotransferase, putative [Ricinus communis] |
| 12 |
Hb_000343_070 |
0.0765525181 |
- |
- |
hypothetical protein JCGZ_06265 [Jatropha curcas] |
| 13 |
Hb_000232_040 |
0.0770709976 |
transcription factor |
TF Family: NAC |
PREDICTED: NAC domain-containing protein 86 isoform X2 [Populus euphratica] |
| 14 |
Hb_001048_010 |
0.0787314598 |
- |
- |
PREDICTED: probable xyloglucan endotransglucosylase/hydrolase protein 7 [Jatropha curcas] |
| 15 |
Hb_002809_180 |
0.0787635473 |
- |
- |
PREDICTED: homeobox protein HD1 [Jatropha curcas] |
| 16 |
Hb_061256_020 |
0.0810916115 |
- |
- |
PREDICTED: UPF0392 protein RCOM_0530710 [Jatropha curcas] |
| 17 |
Hb_002062_010 |
0.0812294243 |
- |
- |
hypothetical protein JCGZ_02762 [Jatropha curcas] |
| 18 |
Hb_010589_010 |
0.0817811349 |
- |
- |
PREDICTED: classical arabinogalactan protein 9-like [Jatropha curcas] |
| 19 |
Hb_000485_100 |
0.0829662142 |
- |
- |
PREDICTED: putative UPF0481 protein At3g02645 [Populus euphratica] |
| 20 |
Hb_010172_040 |
0.0835933077 |
- |
- |
PREDICTED: BAG family molecular chaperone regulator 3-like isoform X1 [Jatropha curcas] |