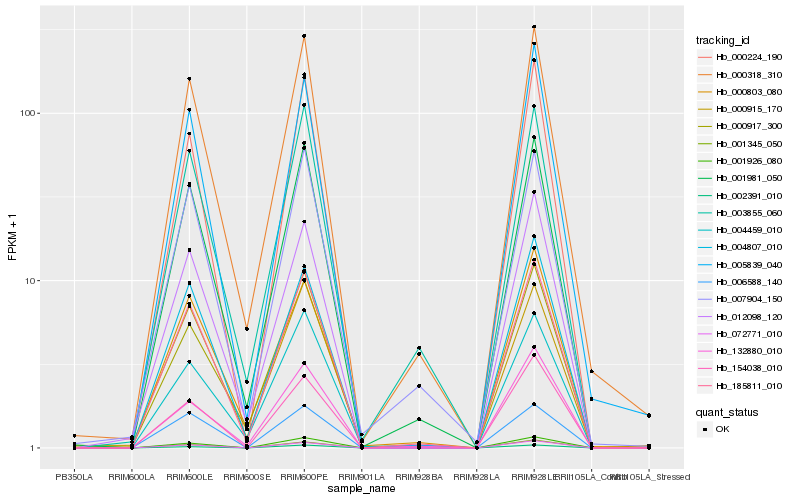
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_185811_010 |
0.0 |
- |
- |
Flavonol 4'-sulfotransferase, putative [Ricinus communis] |
| 2 |
Hb_001981_050 |
0.0227562432 |
- |
- |
PREDICTED: aldehyde dehydrogenase family 3 member F1-like [Jatropha curcas] |
| 3 |
Hb_000318_310 |
0.0475701783 |
- |
- |
hypothetical protein POPTR_0009s11190g [Populus trichocarpa] |
| 4 |
Hb_001345_050 |
0.0616337071 |
- |
- |
hypothetical protein POPTR_0014s14480g [Populus trichocarpa] |
| 5 |
Hb_072771_010 |
0.0622710618 |
- |
- |
Endo-1,4-beta-xylanase C precursor, putative [Ricinus communis] |
| 6 |
Hb_004807_010 |
0.0624751568 |
- |
- |
PREDICTED: auxin-induced protein 15A-like [Jatropha curcas] |
| 7 |
Hb_003855_060 |
0.0633634569 |
- |
- |
PREDICTED: uncharacterized protein At4g06744-like [Jatropha curcas] |
| 8 |
Hb_001926_080 |
0.0680893282 |
- |
- |
PREDICTED: uncharacterized protein LOC103454919 [Malus domestica] |
| 9 |
Hb_012098_120 |
0.0683811786 |
- |
- |
PREDICTED: putative elongation of fatty acids protein DDB_G0272012 [Jatropha curcas] |
| 10 |
Hb_000917_300 |
0.0700094509 |
transcription factor |
TF Family: bHLH |
PREDICTED: transcription factor bHLH30-like [Populus euphratica] |
| 11 |
Hb_002391_010 |
0.072850429 |
- |
- |
PREDICTED: probable acyl-activating enzyme 1, peroxisomal [Jatropha curcas] |
| 12 |
Hb_007904_150 |
0.0732801535 |
- |
- |
PREDICTED: probable pectate lyase 5 [Populus euphratica] |
| 13 |
Hb_000224_190 |
0.0755234993 |
- |
- |
zinc finger protein, putative [Ricinus communis] |
| 14 |
Hb_004459_010 |
0.0763166138 |
- |
- |
PREDICTED: phospholipase A1-IIgamma-like [Jatropha curcas] |
| 15 |
Hb_000915_170 |
0.0794076529 |
transcription factor |
TF Family: NAC |
PREDICTED: NAC domain-containing protein 7 isoform X1 [Vitis vinifera] |
| 16 |
Hb_000803_080 |
0.0794677744 |
desease resistance |
Gene Name: ABC_tran |
ATP-binding cassette transporter, putative [Ricinus communis] |
| 17 |
Hb_005839_040 |
0.0798725451 |
- |
- |
hypothetical protein POPTR_0019s08420g [Populus trichocarpa] |
| 18 |
Hb_006588_140 |
0.0823338967 |
- |
- |
PREDICTED: uncharacterized protein LOC104906772 [Beta vulgaris subsp. vulgaris] |
| 19 |
Hb_154038_010 |
0.0826793222 |
- |
- |
cytochrome P450, putative [Ricinus communis] |
| 20 |
Hb_132880_010 |
0.08406949 |
- |
- |
PREDICTED: uncharacterized protein LOC105641595 [Jatropha curcas] |