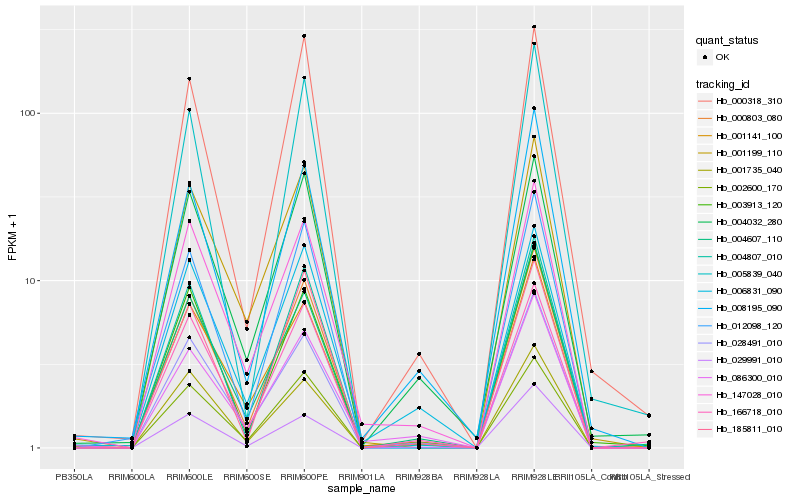
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000803_080 |
0.0 |
desease resistance |
Gene Name: ABC_tran |
ATP-binding cassette transporter, putative [Ricinus communis] |
| 2 |
Hb_003913_120 |
0.0561295675 |
transcription factor |
TF Family: MYB |
hypothetical protein POPTR_0015s09430g [Populus trichocarpa] |
| 3 |
Hb_004807_010 |
0.0628365595 |
- |
- |
PREDICTED: auxin-induced protein 15A-like [Jatropha curcas] |
| 4 |
Hb_028491_010 |
0.0642795947 |
- |
- |
hypothetical protein JCGZ_12857 [Jatropha curcas] |
| 5 |
Hb_004607_110 |
0.0654859463 |
- |
- |
PREDICTED: serine/threonine-protein kinase Nek6 [Jatropha curcas] |
| 6 |
Hb_147028_010 |
0.0659922802 |
transcription factor |
TF Family: HB |
PREDICTED: homeobox protein HD1 [Prunus mume] |
| 7 |
Hb_012098_120 |
0.0690582329 |
- |
- |
PREDICTED: putative elongation of fatty acids protein DDB_G0272012 [Jatropha curcas] |
| 8 |
Hb_166718_010 |
0.0692023704 |
- |
- |
heat-shock protein, putative [Ricinus communis] |
| 9 |
Hb_001199_110 |
0.0736483254 |
- |
- |
PREDICTED: uncharacterized protein ycf36 isoform X1 [Jatropha curcas] |
| 10 |
Hb_001141_100 |
0.0762622903 |
- |
- |
PREDICTED: type I inositol 1,4,5-trisphosphate 5-phosphatase CVP2-like [Jatropha curcas] |
| 11 |
Hb_000318_310 |
0.0792765825 |
- |
- |
hypothetical protein POPTR_0009s11190g [Populus trichocarpa] |
| 12 |
Hb_185811_010 |
0.0794677744 |
- |
- |
Flavonol 4'-sulfotransferase, putative [Ricinus communis] |
| 13 |
Hb_005839_040 |
0.0808737148 |
- |
- |
hypothetical protein POPTR_0019s08420g [Populus trichocarpa] |
| 14 |
Hb_008195_090 |
0.0818855076 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 15 |
Hb_002600_170 |
0.0824358286 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 16 |
Hb_029991_010 |
0.0832737999 |
- |
- |
hypothetical protein PRUPE_ppa001615mg [Prunus persica] |
| 17 |
Hb_006831_090 |
0.0833703436 |
- |
- |
PREDICTED: uncharacterized protein LOC105642125 isoform X1 [Jatropha curcas] |
| 18 |
Hb_086300_010 |
0.0841642203 |
- |
- |
hydrolase, hydrolyzing O-glycosyl compounds, putative [Ricinus communis] |
| 19 |
Hb_001735_040 |
0.0845900436 |
- |
- |
PREDICTED: F-box protein SKIP2-like isoform X2 [Populus euphratica] |
| 20 |
Hb_004032_280 |
0.0849055957 |
- |
- |
PREDICTED: uncharacterized protein LOC105635149 [Jatropha curcas] |