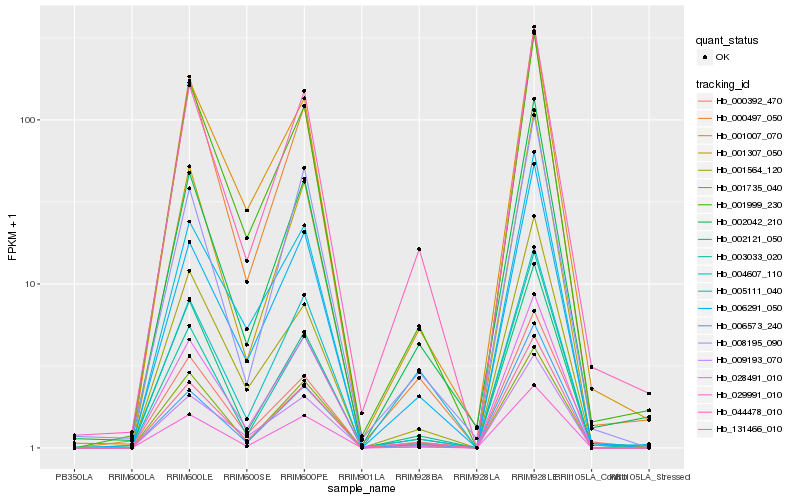
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_131466_010 |
0.0 |
- |
- |
PREDICTED: putative amidase C869.01 [Jatropha curcas] |
| 2 |
Hb_029991_010 |
0.0460599131 |
- |
- |
hypothetical protein PRUPE_ppa001615mg [Prunus persica] |
| 3 |
Hb_006291_050 |
0.0516665117 |
- |
- |
PREDICTED: polyribonucleotide nucleotidyltransferase 2, mitochondrial isoform X1 [Jatropha curcas] |
| 4 |
Hb_009193_070 |
0.0629268162 |
- |
- |
fatty acid elongase 3-ketoacyl-CoA synthase 1 [Arabidopsis thaliana] |
| 5 |
Hb_004607_110 |
0.0633193746 |
- |
- |
PREDICTED: serine/threonine-protein kinase Nek6 [Jatropha curcas] |
| 6 |
Hb_002042_210 |
0.0644364076 |
- |
- |
PREDICTED: UPF0603 protein At1g54780, chloroplastic [Jatropha curcas] |
| 7 |
Hb_028491_010 |
0.0648090473 |
- |
- |
hypothetical protein JCGZ_12857 [Jatropha curcas] |
| 8 |
Hb_044478_010 |
0.0683129318 |
- |
- |
plastoquinol-plastocyanin reductase, putative [Ricinus communis] |
| 9 |
Hb_005111_040 |
0.0686299251 |
- |
- |
glucose-6-phosphate dehydrogenase [Hevea brasiliensis] |
| 10 |
Hb_001999_230 |
0.0686847013 |
- |
- |
PREDICTED: uncharacterized protein LOC105630678 [Jatropha curcas] |
| 11 |
Hb_006573_240 |
0.0763103014 |
- |
- |
PREDICTED: anthocyanidin 5,3-O-glucosyltransferase-like [Jatropha curcas] |
| 12 |
Hb_003033_020 |
0.0767439212 |
- |
- |
PREDICTED: glucomannan 4-beta-mannosyltransferase 9-like [Jatropha curcas] |
| 13 |
Hb_000497_050 |
0.0783884247 |
- |
- |
PREDICTED: photosystem II repair protein PSB27-H1, chloroplastic [Jatropha curcas] |
| 14 |
Hb_008195_090 |
0.0787169282 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 15 |
Hb_001307_050 |
0.0787588041 |
- |
- |
PREDICTED: protein NRT1/ PTR FAMILY 5.2-like [Jatropha curcas] |
| 16 |
Hb_001735_040 |
0.079893274 |
- |
- |
PREDICTED: F-box protein SKIP2-like isoform X2 [Populus euphratica] |
| 17 |
Hb_002121_050 |
0.0799426778 |
- |
- |
hypothetical protein EUGRSUZ_D02586 [Eucalyptus grandis] |
| 18 |
Hb_001007_070 |
0.081841707 |
- |
- |
thioredoxin m(mitochondrial)-type, putative [Ricinus communis] |
| 19 |
Hb_001564_120 |
0.0821563521 |
- |
- |
PREDICTED: peptidyl-prolyl cis-trans isomerase CYP26-2, chloroplastic [Populus euphratica] |
| 20 |
Hb_000392_470 |
0.0822471814 |
- |
- |
conserved hypothetical protein [Ricinus communis] |