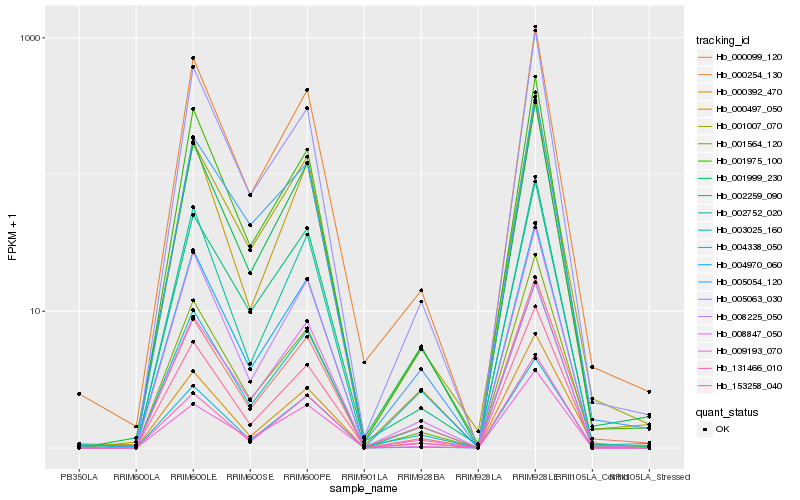
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001999_230 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105630678 [Jatropha curcas] |
| 2 |
Hb_001007_070 |
0.0375228363 |
- |
- |
thioredoxin m(mitochondrial)-type, putative [Ricinus communis] |
| 3 |
Hb_000254_130 |
0.043140119 |
- |
- |
Photosystem I reaction center subunit VI, chloroplast precursor, putative [Ricinus communis] |
| 4 |
Hb_000099_120 |
0.0485194159 |
- |
- |
PREDICTED: GDT1-like protein 1, chloroplastic isoform X3 [Jatropha curcas] |
| 5 |
Hb_000497_050 |
0.0521412154 |
- |
- |
PREDICTED: photosystem II repair protein PSB27-H1, chloroplastic [Jatropha curcas] |
| 6 |
Hb_001564_120 |
0.0523524502 |
- |
- |
PREDICTED: peptidyl-prolyl cis-trans isomerase CYP26-2, chloroplastic [Populus euphratica] |
| 7 |
Hb_001975_100 |
0.0544478322 |
- |
- |
chlorophyll a-b binding protein 151, chloroplastic [Jatropha curcas] |
| 8 |
Hb_008225_050 |
0.0551546333 |
- |
- |
hypothetical protein PRUPE_ppa004332mg [Prunus persica] |
| 9 |
Hb_005063_030 |
0.0559540028 |
- |
- |
Photosystem II reaction center W protein, chloroplast precursor, putative [Ricinus communis] |
| 10 |
Hb_004338_050 |
0.0579130182 |
- |
- |
hypothetical protein POPTR_0004s02760g [Populus trichocarpa] |
| 11 |
Hb_003025_160 |
0.0583167178 |
- |
- |
Protein HVA22, putative [Ricinus communis] |
| 12 |
Hb_002752_020 |
0.0601074268 |
- |
- |
FKBP-type peptidyl-prolyl cis-trans isomerase 3 family protein [Populus trichocarpa] |
| 13 |
Hb_009193_070 |
0.0612886199 |
- |
- |
fatty acid elongase 3-ketoacyl-CoA synthase 1 [Arabidopsis thaliana] |
| 14 |
Hb_002259_090 |
0.0634248267 |
- |
- |
PREDICTED: ABC transporter G family member 11 [Jatropha curcas] |
| 15 |
Hb_004970_060 |
0.0653230038 |
- |
- |
PREDICTED: uncharacterized protein LOC100259061 [Vitis vinifera] |
| 16 |
Hb_008847_050 |
0.0671232379 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_131466_010 |
0.0686847013 |
- |
- |
PREDICTED: putative amidase C869.01 [Jatropha curcas] |
| 18 |
Hb_005054_120 |
0.0688559678 |
- |
- |
PREDICTED: sedoheptulose-1,7-bisphosphatase, chloroplastic [Jatropha curcas] |
| 19 |
Hb_153258_040 |
0.0699793992 |
- |
- |
PREDICTED: type I inositol 1,4,5-trisphosphate 5-phosphatase CVP2-like [Jatropha curcas] |
| 20 |
Hb_000392_470 |
0.0704346238 |
- |
- |
conserved hypothetical protein [Ricinus communis] |