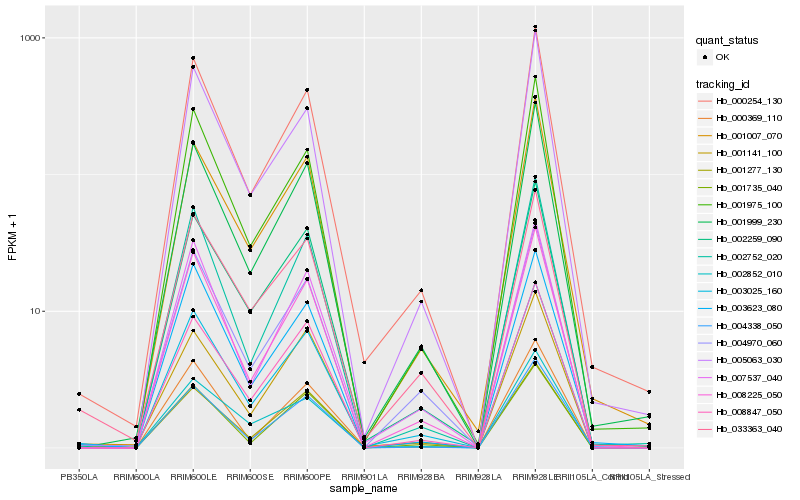
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_003025_160 |
0.0 |
- |
- |
Protein HVA22, putative [Ricinus communis] |
| 2 |
Hb_008225_050 |
0.0460102787 |
- |
- |
hypothetical protein PRUPE_ppa004332mg [Prunus persica] |
| 3 |
Hb_000254_130 |
0.0465196387 |
- |
- |
Photosystem I reaction center subunit VI, chloroplast precursor, putative [Ricinus communis] |
| 4 |
Hb_004970_060 |
0.0514669728 |
- |
- |
PREDICTED: uncharacterized protein LOC100259061 [Vitis vinifera] |
| 5 |
Hb_002259_090 |
0.0563942675 |
- |
- |
PREDICTED: ABC transporter G family member 11 [Jatropha curcas] |
| 6 |
Hb_007537_040 |
0.0574944631 |
- |
- |
PREDICTED: uncharacterized protein LOC105803495 [Gossypium raimondii] |
| 7 |
Hb_001999_230 |
0.0583167178 |
- |
- |
PREDICTED: uncharacterized protein LOC105630678 [Jatropha curcas] |
| 8 |
Hb_008847_050 |
0.0621658894 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 9 |
Hb_000369_110 |
0.0654312156 |
- |
- |
ankyrin repeat-containing protein, putative [Ricinus communis] |
| 10 |
Hb_033363_040 |
0.0659806272 |
- |
- |
PREDICTED: probable galacturonosyltransferase-like 3 [Jatropha curcas] |
| 11 |
Hb_001975_100 |
0.0680294892 |
- |
- |
chlorophyll a-b binding protein 151, chloroplastic [Jatropha curcas] |
| 12 |
Hb_002752_020 |
0.0688705078 |
- |
- |
FKBP-type peptidyl-prolyl cis-trans isomerase 3 family protein [Populus trichocarpa] |
| 13 |
Hb_001007_070 |
0.0692357489 |
- |
- |
thioredoxin m(mitochondrial)-type, putative [Ricinus communis] |
| 14 |
Hb_005063_030 |
0.0750071996 |
- |
- |
Photosystem II reaction center W protein, chloroplast precursor, putative [Ricinus communis] |
| 15 |
Hb_001141_100 |
0.0758804092 |
- |
- |
PREDICTED: type I inositol 1,4,5-trisphosphate 5-phosphatase CVP2-like [Jatropha curcas] |
| 16 |
Hb_004338_050 |
0.0769444062 |
- |
- |
hypothetical protein POPTR_0004s02760g [Populus trichocarpa] |
| 17 |
Hb_003623_080 |
0.0774127378 |
- |
- |
PREDICTED: pheophytinase, chloroplastic [Populus euphratica] |
| 18 |
Hb_001735_040 |
0.0781568714 |
- |
- |
PREDICTED: F-box protein SKIP2-like isoform X2 [Populus euphratica] |
| 19 |
Hb_002852_010 |
0.0781862372 |
- |
- |
leucine-rich repeat receptor protein kinase exs precursor, putative [Ricinus communis] |
| 20 |
Hb_001277_130 |
0.0789966157 |
- |
- |
PREDICTED: dentin sialophosphoprotein-like [Jatropha curcas] |