| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
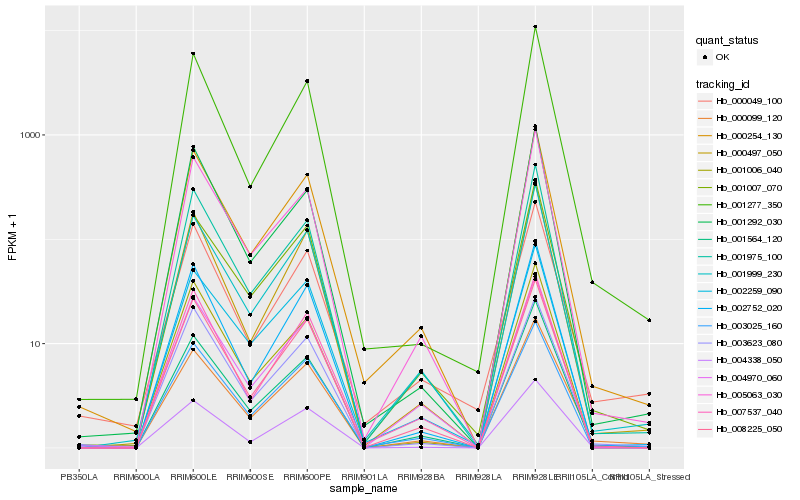
Hb_000254_130 |
0.0 |
- |
- |
Photosystem I reaction center subunit VI, chloroplast precursor, putative [Ricinus communis] |
| 2 |
Hb_001975_100 |
0.0418622104 |
- |
- |
chlorophyll a-b binding protein 151, chloroplastic [Jatropha curcas] |
| 3 |
Hb_001999_230 |
0.043140119 |
- |
- |
PREDICTED: uncharacterized protein LOC105630678 [Jatropha curcas] |
| 4 |
Hb_008225_050 |
0.0457133419 |
- |
- |
hypothetical protein PRUPE_ppa004332mg [Prunus persica] |
| 5 |
Hb_003025_160 |
0.0465196387 |
- |
- |
Protein HVA22, putative [Ricinus communis] |
| 6 |
Hb_005063_030 |
0.048938512 |
- |
- |
Photosystem II reaction center W protein, chloroplast precursor, putative [Ricinus communis] |
| 7 |
Hb_002752_020 |
0.0558861191 |
- |
- |
FKBP-type peptidyl-prolyl cis-trans isomerase 3 family protein [Populus trichocarpa] |
| 8 |
Hb_001007_070 |
0.0597790973 |
- |
- |
thioredoxin m(mitochondrial)-type, putative [Ricinus communis] |
| 9 |
Hb_000497_050 |
0.0605632721 |
- |
- |
PREDICTED: photosystem II repair protein PSB27-H1, chloroplastic [Jatropha curcas] |
| 10 |
Hb_004970_060 |
0.0621095615 |
- |
- |
PREDICTED: uncharacterized protein LOC100259061 [Vitis vinifera] |
| 11 |
Hb_007537_040 |
0.0641717472 |
- |
- |
PREDICTED: uncharacterized protein LOC105803495 [Gossypium raimondii] |
| 12 |
Hb_001277_350 |
0.065973911 |
- |
- |
early light-induced protein, putative [Ricinus communis] |
| 13 |
Hb_000099_120 |
0.0661906895 |
- |
- |
PREDICTED: GDT1-like protein 1, chloroplastic isoform X3 [Jatropha curcas] |
| 14 |
Hb_004338_050 |
0.0663627349 |
- |
- |
hypothetical protein POPTR_0004s02760g [Populus trichocarpa] |
| 15 |
Hb_003623_080 |
0.066996677 |
- |
- |
PREDICTED: pheophytinase, chloroplastic [Populus euphratica] |
| 16 |
Hb_001564_120 |
0.0679643087 |
- |
- |
PREDICTED: peptidyl-prolyl cis-trans isomerase CYP26-2, chloroplastic [Populus euphratica] |
| 17 |
Hb_002259_090 |
0.0688448361 |
- |
- |
PREDICTED: ABC transporter G family member 11 [Jatropha curcas] |
| 18 |
Hb_001006_040 |
0.0696131633 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At5g13770, chloroplastic-like isoform X1 [Populus euphratica] |
| 19 |
Hb_000049_100 |
0.070423475 |
- |
- |
Photosystem II reaction center W protein, putative [Ricinus communis] |
| 20 |
Hb_001292_030 |
0.0723494355 |
- |
- |
PREDICTED: chlorophyll a-b binding protein CP26, chloroplastic [Jatropha curcas] |