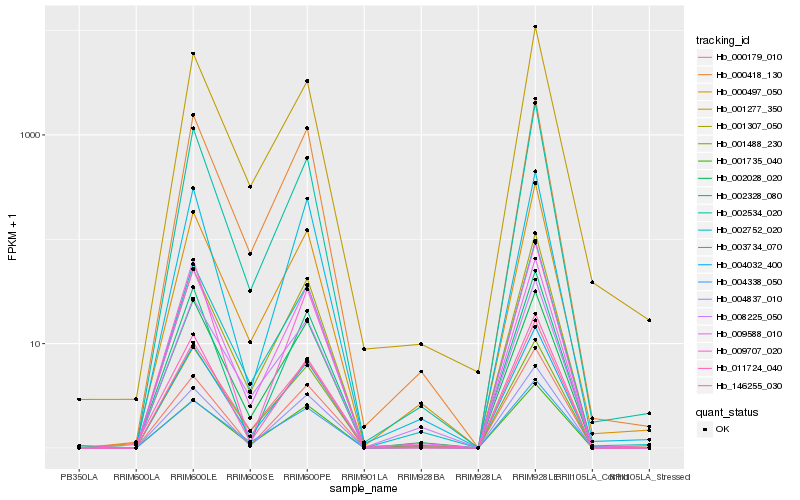
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_004032_400 |
0.0 |
- |
- |
PREDICTED: uncharacterized transporter YBR287W-like [Jatropha curcas] |
| 2 |
Hb_146255_030 |
0.0306841806 |
transcription factor |
TF Family: bZIP |
PREDICTED: transcription factor HY5-like [Jatropha curcas] |
| 3 |
Hb_000418_130 |
0.0405129699 |
- |
- |
PREDICTED: photosystem I reaction center subunit psaK, chloroplastic [Jatropha curcas] |
| 4 |
Hb_002752_020 |
0.0431913534 |
- |
- |
FKBP-type peptidyl-prolyl cis-trans isomerase 3 family protein [Populus trichocarpa] |
| 5 |
Hb_004837_010 |
0.0466888574 |
- |
- |
PREDICTED: aldehyde dehydrogenase family 3 member F1-like [Jatropha curcas] |
| 6 |
Hb_009588_010 |
0.048320209 |
- |
- |
sigma factor sigb regulation protein rsbq, putative [Ricinus communis] |
| 7 |
Hb_002028_020 |
0.053879282 |
- |
- |
acyltransferase, putative [Ricinus communis] |
| 8 |
Hb_000497_050 |
0.0566009331 |
- |
- |
PREDICTED: photosystem II repair protein PSB27-H1, chloroplastic [Jatropha curcas] |
| 9 |
Hb_002534_020 |
0.057807504 |
- |
- |
PREDICTED: chlorophyll a-b binding protein 4, chloroplastic [Gossypium raimondii] |
| 10 |
Hb_001488_230 |
0.0599427083 |
- |
- |
PREDICTED: serine carboxypeptidase-like 25 isoform X2 [Populus euphratica] |
| 11 |
Hb_002328_080 |
0.0605101678 |
- |
- |
short-chain dehydrogenase, putative [Ricinus communis] |
| 12 |
Hb_000179_010 |
0.0618515284 |
- |
- |
PREDICTED: alkane hydroxylase MAH1-like [Jatropha curcas] |
| 13 |
Hb_003734_070 |
0.0630011116 |
- |
- |
chlorophyll A/B binding protein, putative [Ricinus communis] |
| 14 |
Hb_001307_050 |
0.064876085 |
- |
- |
PREDICTED: protein NRT1/ PTR FAMILY 5.2-like [Jatropha curcas] |
| 15 |
Hb_004338_050 |
0.0666614057 |
- |
- |
hypothetical protein POPTR_0004s02760g [Populus trichocarpa] |
| 16 |
Hb_001735_040 |
0.0682514886 |
- |
- |
PREDICTED: F-box protein SKIP2-like isoform X2 [Populus euphratica] |
| 17 |
Hb_001277_350 |
0.0706861803 |
- |
- |
early light-induced protein, putative [Ricinus communis] |
| 18 |
Hb_011724_040 |
0.0712260957 |
- |
- |
PREDICTED: uncharacterized protein LOC105632860 [Jatropha curcas] |
| 19 |
Hb_009707_020 |
0.0717304632 |
- |
- |
PREDICTED: 3-ketoacyl-CoA synthase 12 [Jatropha curcas] |
| 20 |
Hb_008225_050 |
0.0718019261 |
- |
- |
hypothetical protein PRUPE_ppa004332mg [Prunus persica] |