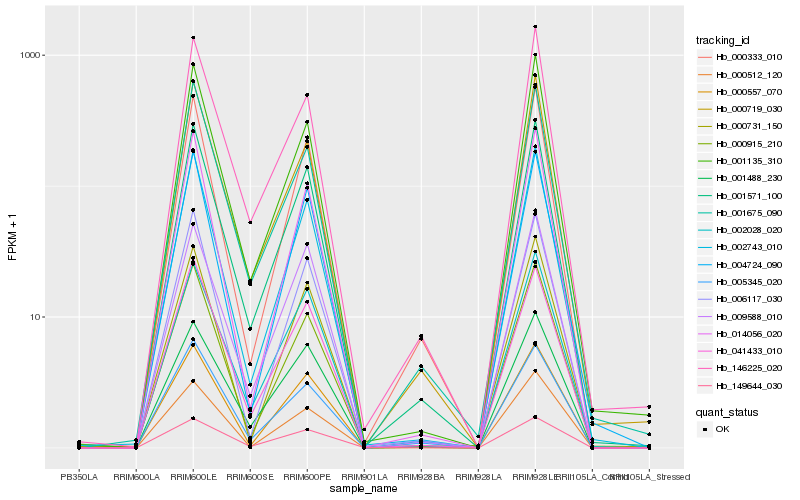
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001571_100 |
0.0 |
- |
- |
chlorophyll A-B binding family protein [Populus trichocarpa] |
| 2 |
Hb_002743_010 |
0.0392095147 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_002028_020 |
0.04484509 |
- |
- |
acyltransferase, putative [Ricinus communis] |
| 4 |
Hb_005345_020 |
0.0457194005 |
- |
- |
Root phototropism protein, putative [Ricinus communis] |
| 5 |
Hb_000333_010 |
0.0479591225 |
- |
- |
hypothetical protein POPTR_0006s14510g [Populus trichocarpa] |
| 6 |
Hb_041433_010 |
0.0480202789 |
transcription factor |
TF Family: G2-like |
DNA binding protein, putative [Ricinus communis] |
| 7 |
Hb_000557_070 |
0.0484536424 |
- |
- |
S-locus-specific glycoprotein S13 precursor, putative [Ricinus communis] |
| 8 |
Hb_000719_030 |
0.0512436505 |
- |
- |
Photosystem I reaction center subunit III, chloroplast precursor, putative [Ricinus communis] |
| 9 |
Hb_000915_210 |
0.0532810263 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 10 |
Hb_149644_030 |
0.0567198247 |
- |
- |
hypothetical protein POPTR_0005s18750g [Populus trichocarpa] |
| 11 |
Hb_014056_020 |
0.0569374589 |
- |
- |
zinc finger protein, putative [Ricinus communis] |
| 12 |
Hb_000512_120 |
0.057357143 |
transcription factor |
TF Family: B3 |
PREDICTED: B3 domain-containing transcription factor NGA1-like [Jatropha curcas] |
| 13 |
Hb_001135_310 |
0.0579946965 |
- |
- |
Chlorophyll a-b binding protein 6A [Morus notabilis] |
| 14 |
Hb_146225_020 |
0.058479011 |
- |
- |
chlorophyll A/B binding protein, putative [Ricinus communis] |
| 15 |
Hb_006117_030 |
0.0594623578 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 16 |
Hb_001675_090 |
0.0600034359 |
- |
- |
Photosystem I reaction center subunit VI, chloroplast precursor, putative [Ricinus communis] |
| 17 |
Hb_001488_230 |
0.0604436906 |
- |
- |
PREDICTED: serine carboxypeptidase-like 25 isoform X2 [Populus euphratica] |
| 18 |
Hb_004724_090 |
0.0630619934 |
- |
- |
PREDICTED: uncharacterized protein LOC105638638 [Jatropha curcas] |
| 19 |
Hb_009588_010 |
0.0642240236 |
- |
- |
sigma factor sigb regulation protein rsbq, putative [Ricinus communis] |
| 20 |
Hb_000731_150 |
0.0649596243 |
- |
- |
PREDICTED: cytochrome P450 77A1 [Jatropha curcas] |