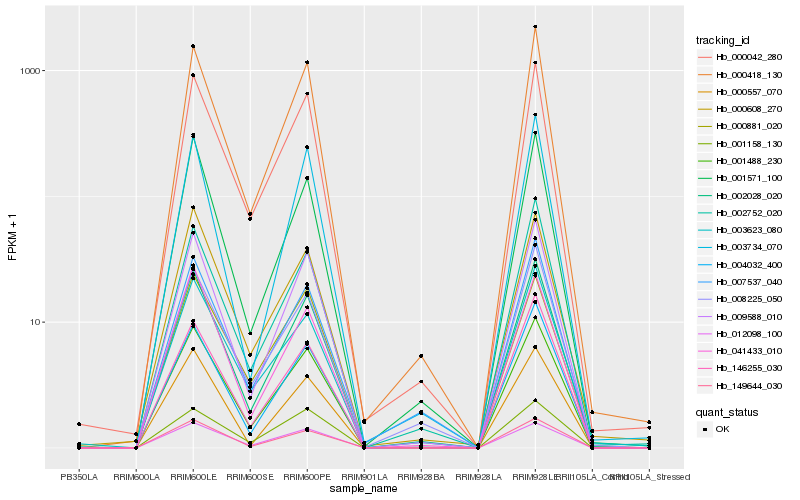
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001488_230 |
0.0 |
- |
- |
PREDICTED: serine carboxypeptidase-like 25 isoform X2 [Populus euphratica] |
| 2 |
Hb_002028_020 |
0.029221489 |
- |
- |
acyltransferase, putative [Ricinus communis] |
| 3 |
Hb_000042_280 |
0.0316274956 |
- |
- |
chlorophyll A/B binding protein, putative [Ricinus communis] |
| 4 |
Hb_009588_010 |
0.0367163109 |
- |
- |
sigma factor sigb regulation protein rsbq, putative [Ricinus communis] |
| 5 |
Hb_000418_130 |
0.0408165954 |
- |
- |
PREDICTED: photosystem I reaction center subunit psaK, chloroplastic [Jatropha curcas] |
| 6 |
Hb_149644_030 |
0.0559896544 |
- |
- |
hypothetical protein POPTR_0005s18750g [Populus trichocarpa] |
| 7 |
Hb_004032_400 |
0.0599427083 |
- |
- |
PREDICTED: uncharacterized transporter YBR287W-like [Jatropha curcas] |
| 8 |
Hb_001571_100 |
0.0604436906 |
- |
- |
chlorophyll A-B binding family protein [Populus trichocarpa] |
| 9 |
Hb_012098_100 |
0.0612438602 |
- |
- |
zinc finger protein, putative [Ricinus communis] |
| 10 |
Hb_003623_080 |
0.0695798245 |
- |
- |
PREDICTED: pheophytinase, chloroplastic [Populus euphratica] |
| 11 |
Hb_000608_270 |
0.0725892983 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_146255_030 |
0.0735343582 |
transcription factor |
TF Family: bZIP |
PREDICTED: transcription factor HY5-like [Jatropha curcas] |
| 13 |
Hb_007537_040 |
0.0741290535 |
- |
- |
PREDICTED: uncharacterized protein LOC105803495 [Gossypium raimondii] |
| 14 |
Hb_041433_010 |
0.0752367142 |
transcription factor |
TF Family: G2-like |
DNA binding protein, putative [Ricinus communis] |
| 15 |
Hb_008225_050 |
0.0753429891 |
- |
- |
hypothetical protein PRUPE_ppa004332mg [Prunus persica] |
| 16 |
Hb_002752_020 |
0.0757063615 |
- |
- |
FKBP-type peptidyl-prolyl cis-trans isomerase 3 family protein [Populus trichocarpa] |
| 17 |
Hb_000557_070 |
0.0772925587 |
- |
- |
S-locus-specific glycoprotein S13 precursor, putative [Ricinus communis] |
| 18 |
Hb_001158_130 |
0.0826257661 |
- |
- |
PREDICTED: protein TAPETUM DETERMINANT 1-like [Jatropha curcas] |
| 19 |
Hb_000881_020 |
0.0828954735 |
- |
- |
hypothetical protein MIMGU_mgv1a022110mg, partial [Erythranthe guttata] |
| 20 |
Hb_003734_070 |
0.0845903081 |
- |
- |
chlorophyll A/B binding protein, putative [Ricinus communis] |