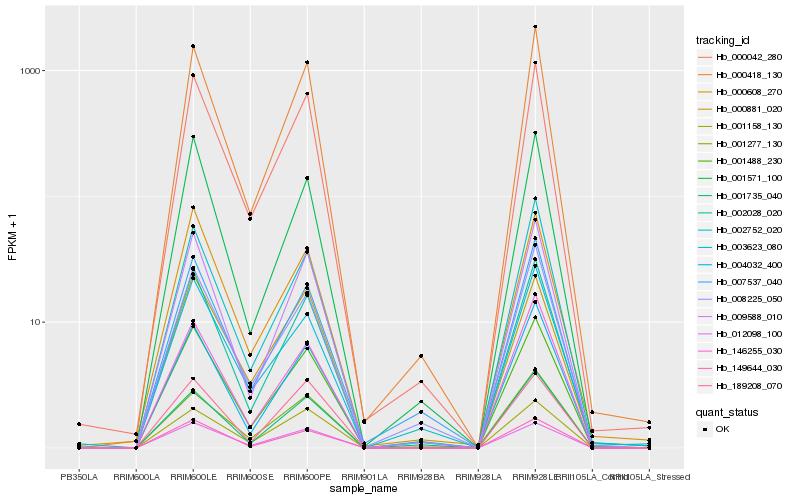
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000042_280 |
0.0 |
- |
- |
chlorophyll A/B binding protein, putative [Ricinus communis] |
| 2 |
Hb_001488_230 |
0.0316274956 |
- |
- |
PREDICTED: serine carboxypeptidase-like 25 isoform X2 [Populus euphratica] |
| 3 |
Hb_000418_130 |
0.0389631856 |
- |
- |
PREDICTED: photosystem I reaction center subunit psaK, chloroplastic [Jatropha curcas] |
| 4 |
Hb_002028_020 |
0.046859861 |
- |
- |
acyltransferase, putative [Ricinus communis] |
| 5 |
Hb_009588_010 |
0.0498937006 |
- |
- |
sigma factor sigb regulation protein rsbq, putative [Ricinus communis] |
| 6 |
Hb_012098_100 |
0.0578016738 |
- |
- |
zinc finger protein, putative [Ricinus communis] |
| 7 |
Hb_149644_030 |
0.0657059781 |
- |
- |
hypothetical protein POPTR_0005s18750g [Populus trichocarpa] |
| 8 |
Hb_001158_130 |
0.0658211617 |
- |
- |
PREDICTED: protein TAPETUM DETERMINANT 1-like [Jatropha curcas] |
| 9 |
Hb_004032_400 |
0.0718491373 |
- |
- |
PREDICTED: uncharacterized transporter YBR287W-like [Jatropha curcas] |
| 10 |
Hb_000881_020 |
0.072532652 |
- |
- |
hypothetical protein MIMGU_mgv1a022110mg, partial [Erythranthe guttata] |
| 11 |
Hb_003623_080 |
0.0728130048 |
- |
- |
PREDICTED: pheophytinase, chloroplastic [Populus euphratica] |
| 12 |
Hb_008225_050 |
0.0752291797 |
- |
- |
hypothetical protein PRUPE_ppa004332mg [Prunus persica] |
| 13 |
Hb_007537_040 |
0.0778668522 |
- |
- |
PREDICTED: uncharacterized protein LOC105803495 [Gossypium raimondii] |
| 14 |
Hb_002752_020 |
0.0796676807 |
- |
- |
FKBP-type peptidyl-prolyl cis-trans isomerase 3 family protein [Populus trichocarpa] |
| 15 |
Hb_001277_130 |
0.0799696367 |
- |
- |
PREDICTED: dentin sialophosphoprotein-like [Jatropha curcas] |
| 16 |
Hb_000608_270 |
0.0800812583 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_001735_040 |
0.0824202997 |
- |
- |
PREDICTED: F-box protein SKIP2-like isoform X2 [Populus euphratica] |
| 18 |
Hb_001571_100 |
0.0829665466 |
- |
- |
chlorophyll A-B binding family protein [Populus trichocarpa] |
| 19 |
Hb_189208_070 |
0.0831665344 |
- |
- |
10-deacetylbaccatin III 10-O-acetyltransferase, putative [Ricinus communis] |
| 20 |
Hb_146255_030 |
0.0831977612 |
transcription factor |
TF Family: bZIP |
PREDICTED: transcription factor HY5-like [Jatropha curcas] |