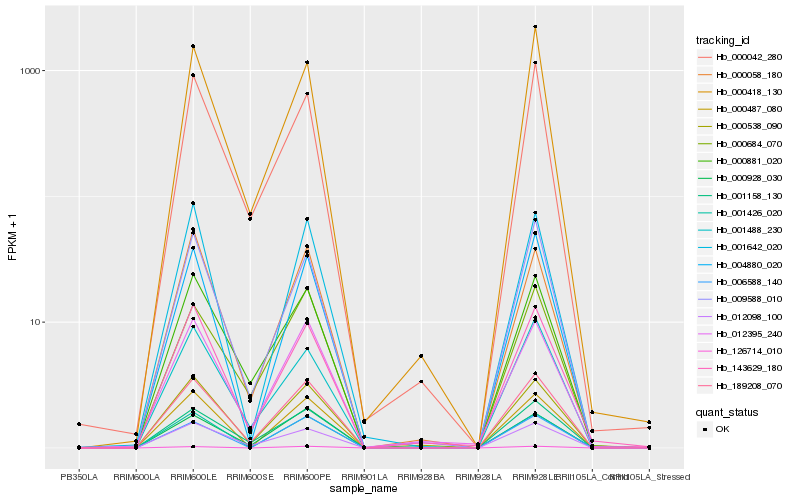
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_189208_070 |
0.0 |
- |
- |
10-deacetylbaccatin III 10-O-acetyltransferase, putative [Ricinus communis] |
| 2 |
Hb_001158_130 |
0.0535013181 |
- |
- |
PREDICTED: protein TAPETUM DETERMINANT 1-like [Jatropha curcas] |
| 3 |
Hb_012098_100 |
0.065809295 |
- |
- |
zinc finger protein, putative [Ricinus communis] |
| 4 |
Hb_000538_090 |
0.0704020194 |
transcription factor |
TF Family: NAC |
hypothetical protein RCOM_1603380 [Ricinus communis] |
| 5 |
Hb_000881_020 |
0.0766443818 |
- |
- |
hypothetical protein MIMGU_mgv1a022110mg, partial [Erythranthe guttata] |
| 6 |
Hb_009588_010 |
0.0784803169 |
- |
- |
sigma factor sigb regulation protein rsbq, putative [Ricinus communis] |
| 7 |
Hb_012395_240 |
0.0803475795 |
- |
- |
PREDICTED: uncharacterized protein LOC105638421 isoform X2 [Jatropha curcas] |
| 8 |
Hb_000928_030 |
0.0819643088 |
transcription factor |
TF Family: bZIP |
PREDICTED: protein ABSCISIC ACID-INSENSITIVE 5 [Jatropha curcas] |
| 9 |
Hb_000042_280 |
0.0831665344 |
- |
- |
chlorophyll A/B binding protein, putative [Ricinus communis] |
| 10 |
Hb_004880_020 |
0.0845957878 |
- |
- |
PREDICTED: G-type lectin S-receptor-like serine/threonine-protein kinase SD1-1 [Jatropha curcas] |
| 11 |
Hb_001426_020 |
0.0853589603 |
- |
- |
hypothetical protein CICLE_v10033594mg, partial [Citrus clementina] |
| 12 |
Hb_000684_070 |
0.0854917263 |
- |
- |
PREDICTED: uncharacterized protein LOC105642198 isoform X1 [Jatropha curcas] |
| 13 |
Hb_143629_180 |
0.0865208248 |
transcription factor |
TF Family: MYB |
myb family transcription factor family protein [Populus trichocarpa] |
| 14 |
Hb_000418_130 |
0.0879801798 |
- |
- |
PREDICTED: photosystem I reaction center subunit psaK, chloroplastic [Jatropha curcas] |
| 15 |
Hb_126714_010 |
0.089772567 |
- |
- |
hypothetical protein CISIN_1g041016mg, partial [Citrus sinensis] |
| 16 |
Hb_000487_080 |
0.0902720572 |
- |
- |
PREDICTED: probable potassium transporter 13 [Jatropha curcas] |
| 17 |
Hb_001488_230 |
0.0903582289 |
- |
- |
PREDICTED: serine carboxypeptidase-like 25 isoform X2 [Populus euphratica] |
| 18 |
Hb_001642_020 |
0.0908471707 |
- |
- |
PREDICTED: probable cinnamyl alcohol dehydrogenase 9 [Jatropha curcas] |
| 19 |
Hb_000058_180 |
0.0913531701 |
- |
- |
PREDICTED: protein ECERIFERUM 3 [Jatropha curcas] |
| 20 |
Hb_006588_140 |
0.091783722 |
- |
- |
PREDICTED: uncharacterized protein LOC104906772 [Beta vulgaris subsp. vulgaris] |