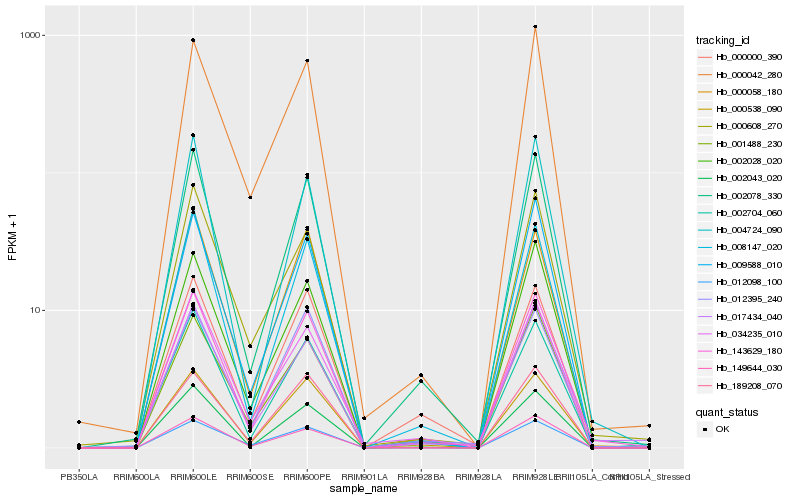
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_143629_180 |
0.0 |
transcription factor |
TF Family: MYB |
myb family transcription factor family protein [Populus trichocarpa] |
| 2 |
Hb_002078_330 |
0.0513120383 |
- |
- |
lipid binding protein, putative [Ricinus communis] |
| 3 |
Hb_002043_020 |
0.0695055248 |
transcription factor |
TF Family: bHLH |
PREDICTED: transcription factor bHLH87 [Jatropha curcas] |
| 4 |
Hb_000538_090 |
0.0698350169 |
transcription factor |
TF Family: NAC |
hypothetical protein RCOM_1603380 [Ricinus communis] |
| 5 |
Hb_017434_040 |
0.0731724178 |
- |
- |
PREDICTED: uncharacterized oxidoreductase At4g09670-like [Jatropha curcas] |
| 6 |
Hb_002704_060 |
0.0746436783 |
- |
- |
PREDICTED: probable inactive leucine-rich repeat receptor-like protein kinase At1g66830 [Jatropha curcas] |
| 7 |
Hb_008147_020 |
0.0754451844 |
- |
- |
hypothetical protein JCGZ_25652 [Jatropha curcas] |
| 8 |
Hb_002028_020 |
0.0799047239 |
- |
- |
acyltransferase, putative [Ricinus communis] |
| 9 |
Hb_009588_010 |
0.0821153467 |
- |
- |
sigma factor sigb regulation protein rsbq, putative [Ricinus communis] |
| 10 |
Hb_012395_240 |
0.0832707122 |
- |
- |
PREDICTED: uncharacterized protein LOC105638421 isoform X2 [Jatropha curcas] |
| 11 |
Hb_000058_180 |
0.0836746689 |
- |
- |
PREDICTED: protein ECERIFERUM 3 [Jatropha curcas] |
| 12 |
Hb_012098_100 |
0.085335289 |
- |
- |
zinc finger protein, putative [Ricinus communis] |
| 13 |
Hb_149644_030 |
0.0858302726 |
- |
- |
hypothetical protein POPTR_0005s18750g [Populus trichocarpa] |
| 14 |
Hb_189208_070 |
0.0865208248 |
- |
- |
10-deacetylbaccatin III 10-O-acetyltransferase, putative [Ricinus communis] |
| 15 |
Hb_004724_090 |
0.0872818533 |
- |
- |
PREDICTED: uncharacterized protein LOC105638638 [Jatropha curcas] |
| 16 |
Hb_001488_230 |
0.0884181764 |
- |
- |
PREDICTED: serine carboxypeptidase-like 25 isoform X2 [Populus euphratica] |
| 17 |
Hb_000608_270 |
0.0887118855 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_000042_280 |
0.0891108311 |
- |
- |
chlorophyll A/B binding protein, putative [Ricinus communis] |
| 19 |
Hb_034235_010 |
0.0905365386 |
- |
- |
PREDICTED: UPF0481 protein At3g47200-like [Jatropha curcas] |
| 20 |
Hb_000000_390 |
0.0907251673 |
- |
- |
PREDICTED: uncharacterized protein LOC105633753 [Jatropha curcas] |