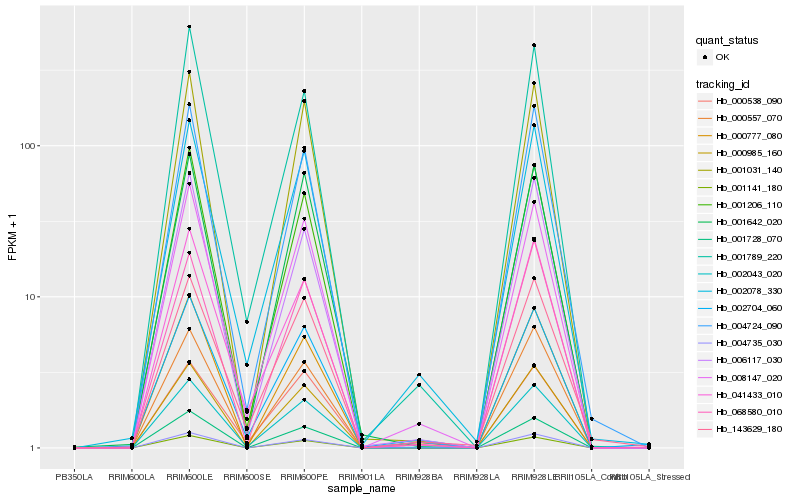
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002043_020 |
0.0 |
transcription factor |
TF Family: bHLH |
PREDICTED: transcription factor bHLH87 [Jatropha curcas] |
| 2 |
Hb_008147_020 |
0.0298950552 |
- |
- |
hypothetical protein JCGZ_25652 [Jatropha curcas] |
| 3 |
Hb_002078_330 |
0.04099468 |
- |
- |
lipid binding protein, putative [Ricinus communis] |
| 4 |
Hb_001206_110 |
0.0490576914 |
- |
- |
PREDICTED: GDSL esterase/lipase At5g45910 [Jatropha curcas] |
| 5 |
Hb_001031_140 |
0.0509612729 |
- |
- |
PREDICTED: GDSL esterase/lipase At5g33370-like [Jatropha curcas] |
| 6 |
Hb_000557_070 |
0.052894129 |
- |
- |
S-locus-specific glycoprotein S13 precursor, putative [Ricinus communis] |
| 7 |
Hb_001141_180 |
0.0539092287 |
- |
- |
non-specific lipid-transfer protein-like protein At2g13820 precursor [Jatropha curcas] |
| 8 |
Hb_004724_090 |
0.055350915 |
- |
- |
PREDICTED: uncharacterized protein LOC105638638 [Jatropha curcas] |
| 9 |
Hb_002704_060 |
0.0584788786 |
- |
- |
PREDICTED: probable inactive leucine-rich repeat receptor-like protein kinase At1g66830 [Jatropha curcas] |
| 10 |
Hb_004735_030 |
0.0609176543 |
- |
- |
PREDICTED: cytochrome P450 734A1 [Jatropha curcas] |
| 11 |
Hb_001728_070 |
0.0616650022 |
- |
- |
PREDICTED: probable glycosyltransferase At3g07620 [Jatropha curcas] |
| 12 |
Hb_000777_080 |
0.0637730454 |
- |
- |
hypothetical protein POPTR_0015s15410g [Populus trichocarpa] |
| 13 |
Hb_041433_010 |
0.0652363336 |
transcription factor |
TF Family: G2-like |
DNA binding protein, putative [Ricinus communis] |
| 14 |
Hb_000985_160 |
0.0675267743 |
- |
- |
PREDICTED: cytochrome P450 724B1-like [Jatropha curcas] |
| 15 |
Hb_001642_020 |
0.0679006255 |
- |
- |
PREDICTED: probable cinnamyl alcohol dehydrogenase 9 [Jatropha curcas] |
| 16 |
Hb_143629_180 |
0.0695055248 |
transcription factor |
TF Family: MYB |
myb family transcription factor family protein [Populus trichocarpa] |
| 17 |
Hb_006117_030 |
0.0695961136 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_068580_010 |
0.0704779395 |
- |
- |
PREDICTED: probable receptor-like serine/threonine-protein kinase At5g57670 isoform X2 [Populus euphratica] |
| 19 |
Hb_001789_220 |
0.0731675444 |
- |
- |
PREDICTED: BAHD acyltransferase DCR-like [Jatropha curcas] |
| 20 |
Hb_000538_090 |
0.0746635572 |
transcription factor |
TF Family: NAC |
hypothetical protein RCOM_1603380 [Ricinus communis] |